I want to read parts from a large (ca. 11 GB) binary file. The currently working solution is to load the entire file ( raw_data ) with fread(), then crop out pieces of interest ( data ).
Question: Is there a faster method of reading small (1-2% of total file, partially sequential reads) parts of a file, given something like a binary mask (i.e. a logical index of specific bytes of interst) in Matlab? Specifics below.
Notes for my specific case:
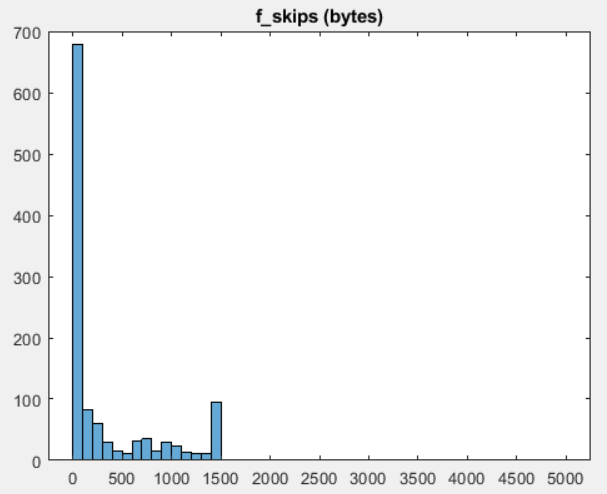
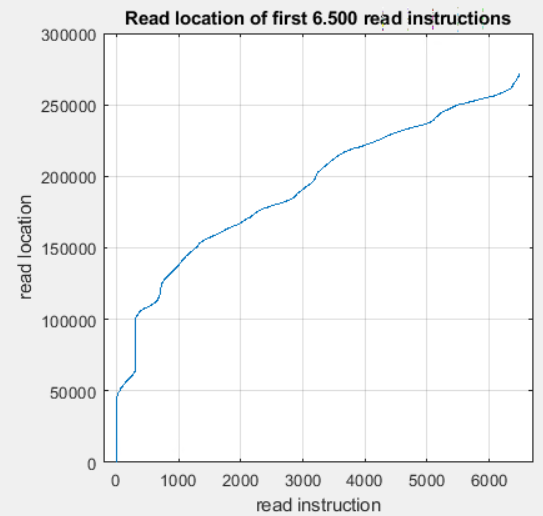
dataof interest (26 e6 bytes, or ca. 24 MB) is roughly 2% ofraw_data(1.2e 10 bytes or ca. 11 GB)- each 600.000 bytes contain ca 6.500 byte reads, which can be broken down to roughly 1.200 read-skip cycles (such as 'read 10 bytes, skip 5000 bytes').
- the read instructions of the total file can be broken down in ca 20.000 similar but (not exactly identical) read-skip cycles (i.e. ca. 20.000x1.200 read-skip cycles)
- The file is read from a GPFS (parallel file system)
- Excessive RAM, newest Matlab ver and all toolboxes are available for the task
My initial idea of fread-fseek cycle proved to be extrodinarily much slower (see psuedocode below) than reading the whole file. Profiling revealed fread() is slowest (being called over a million times probably obvious to the experts here).
Alternatives I considered: memmapfile() [ 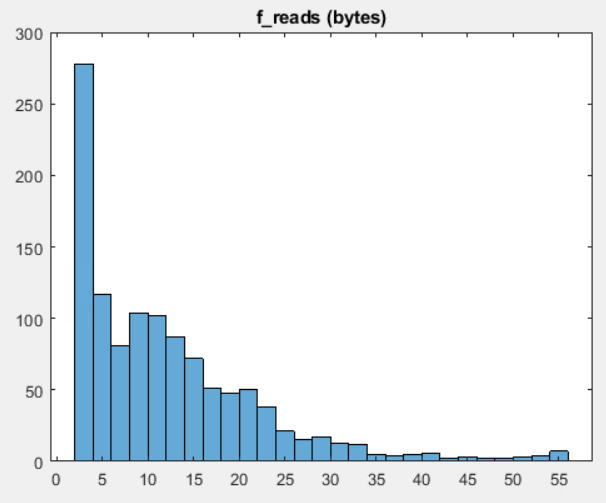
CodePudding user response:
I would do two things to speed up your code:
- preallocate the data array.
- write a C MEX-file to call
freadandfseek.
This is a quick test I did to compare using fread and fseek from MATLAB or C:
%% Create large binary file
data = 1:10000000; % 80 MB
fi = fopen('data.bin', 'wb');
fwrite(fi, data, 'double');
fclose(fi);
n_read = 1;
n_skip = 99;
%% Read using MATLAB
tic
fi = fopen('data.bin', 'rb');
fseek(fi, 0, 'eof');
sz = ftell(fi);
sz = floor(sz / (n_read n_skip));
data = zeros(1, sz);
fseek(fi, 0, 'bof');
for ind = 1:sz
data(ind) = fread(fi, n_read, 'int8');
fseek(fi, n_skip, 'cof');
end
toc
%% Read using C MEX-file
mex fread_test_mex.c
tic
data = fread_test_mex('data.bin', n_read, n_skip);
toc
And this is fread_test_mex.c:
#include <stdio.h>
#include <mex.h>
void mexFunction(int nlhs, mxArray *plhs[],
int nrhs, const mxArray *prhs[])
{
// No testing of inputs...
// inputs = 'data.bin', 1, 99
char* fname = mxArrayToString(prhs[0]);
int n_read = mxGetScalar(prhs[1]);
int n_skip = mxGetScalar(prhs[2]);
FILE* fi = fopen(fname, "rb");
fseek(fi, 0L, SEEK_END);
int sz = ftell(fi);
sz /= n_read n_skip;
plhs[0] = mxCreateNumericMatrix(1, sz, mxDOUBLE_CLASS, mxREAL);
double* data = mxGetPr(plhs[0]);
fseek(fi, 0L, SEEK_SET);
char buffer[1];
for(int ind = 1; ind < sz; ind) {
fread(buffer, 1, n_read, fi);
data[ind] = buffer[0];
fseek(fi, n_skip, SEEK_CUR);
}
fclose(fi);
}
I see this:
Elapsed time is 6.785304 seconds.
Building with 'Xcode with Clang'.
MEX completed successfully.
Elapsed time is 1.376540 seconds.
That is, reading the data is 5x as fast with a C MEX-file. And that time includes loading the MEX-file into memory. A second run is a bit faster (1.14 s) because the MEX-file is already loaded.
In the MATLAB code, if I initialize data = []; and then extend the matrix every time I read like OP does:
tmp = fread(fi, n_read, 'int8');
data = [data, tmp];
then the execution time for that loop was 159 s, with 92.0% of the time spent in the data = [data, tmp] line. Preallocating really is important!
