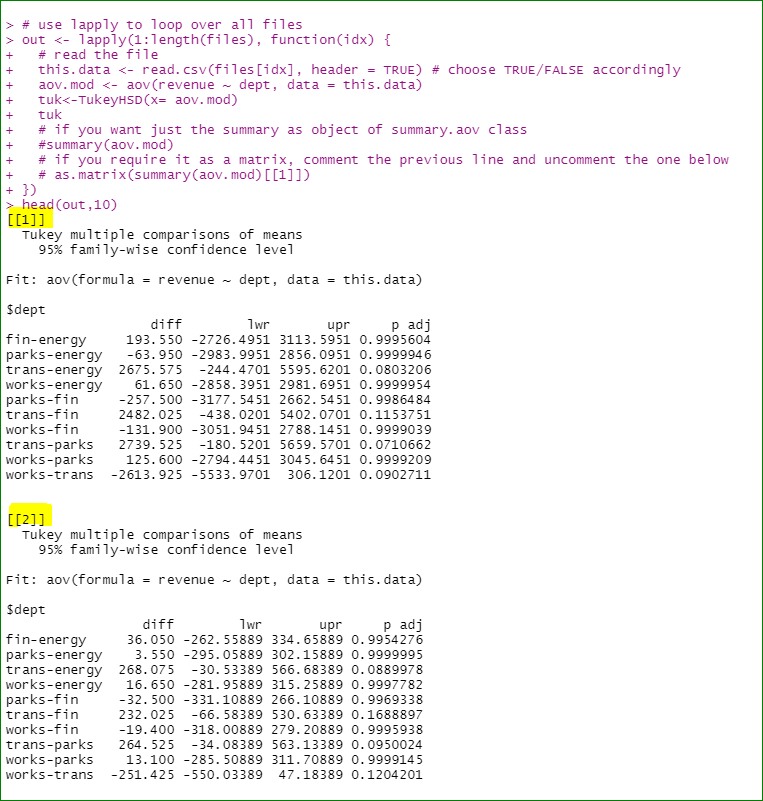
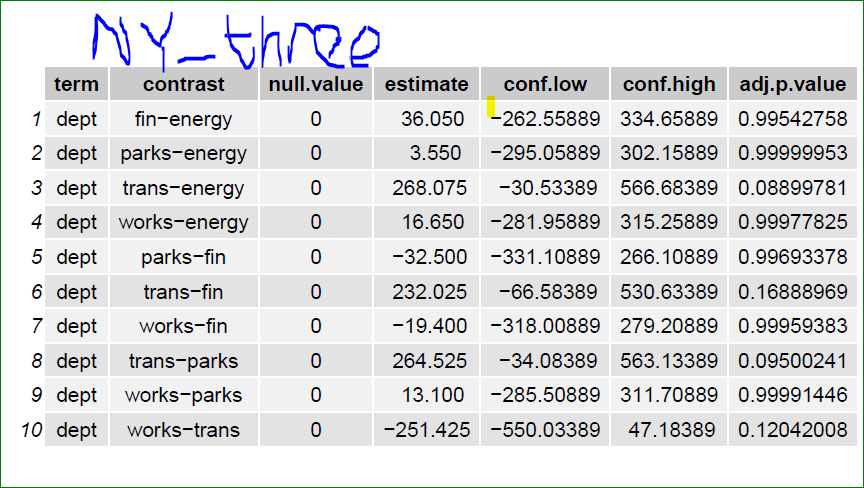
How do I append the name of corresponding file name to the list output of my code. The goal is to be able to trace the outputs in the list to the input csv files. Currently, the list output returns a list index [[1]],[[2]],...,[[5]] (see the snapshot below). I want the corresponding file name included, something like this CA_three , FL_three,...., NY_two
@ Akrun, I want each page to have a corresponding file name
Below there are two codes
Code 1 : code that loops through 5 csv files and returns a list of outputs
[[]](I need help here)Code 2: code to generate 5 csv files used in
Code 1
# Code 1
library(multcompView) # for tukeyHD
# list of 20 csv files in a folder
files <- list.files("C:\\mypath\\", pattern="*.csv", full.names = T)
out <- lapply(1:length(files), function(x) { # use lapply to loop over all files
this_data <- read.csv(files[x], header = TRUE)
aov_mod <- aov(revenue ~ dept, data = this_data)
tuk<-TukeyHSD(x= aov_mod)
tuk
})
out
# Code 2
# In order to generate 5 csv files, copy and paste the code below and save in a new folder
#
state <-c("NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY")
dept <- c("energy","energy","energy","energy","works",'works','works','works','fin','fin','fin','fin','parks','parks','parks','parks','trans','trans','trans','trans')
year <- c("two","two","two","two","two","two","two","two","two","two","two","two","two","two","two","two","two","two","two","two")
revenue <-c(1212.9,1253,1244.4,5123.5,1312,3134,515.8,2449.9,3221.6,3132.5,2235.09,2239.01,3235.01,5223.01,4235.6,2204.5,2315.5,6114,4512,3514.2)
NY_two <-data.frame(state,dept,year,revenue)
# dataset 2
state <-c("NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY","NY")
dept <- c("energy","energy","energy","energy","works",'works','works','works','fin','fin','fin','fin','parks','parks','parks','parks','trans','trans','trans','trans')
year <- rep("five",20)
revenue <-c(1212.9,1253,1244.4,5123.5,1312,3134,515.8,2449.9,3221.6,3132.5,2235.09,2239.01,3235.01,5223.01,4235.6,2204.5,2315.5,6114,4512,3514.2)
NY_five <-data.frame(state,dept,year,revenue)
state <- rep("FL",20)
dept <- c("energy","energy","energy","energy","works",'works','works','works','fin','fin','fin','fin','parks','parks','parks','parks','trans','trans','trans','trans')
year <- rep("three",20)
revenue <-c(112.9,123,124,523.5,112,334,55,449,221.6,332,235,239,235,223,235.6,204,315.5,614,512,514.2)
FL_three <- data.frame(state,dept,year,revenue)
state <- rep("CA",20)
dept <- c("energy","energy","energy","energy","works",'works','works','works','fin','fin','fin','fin','parks','parks','parks','parks','trans','trans','trans','trans')
year <- rep("three",20)
revenue <-c(1102.9,1023,1024,5203.5,1012,3034,505,4049,2021.6,3032,2035,2039,2035,2023,2035.6,2004,3015.5,6014,5012,5014.2)
CA_three <- data.frame(state,dept,year,revenue)
state <- rep("KY",20)
dept <- c("energy","energy","energy","energy","works",'works','works','works','fin','fin','fin','fin','parks','parks','parks','parks','trans','trans','trans','trans')
year <- rep("one",20)
revenue <-c(1102.9,1023,1024,5203.5,1012,3034,505,4049,2021.6,3032,2035,2039,2035,2023,2035.6,2004,3015.5,6014,5012,5014.2)
KY_one <- data.frame(state,dept,year,revenue)
setwd("C:\\define your path\\")
# writing out the file to a newly created folder
write.csv(NY_two,"NY_two.csv",row.names = FALSE)
write.csv(CA_three,"CA_three.csv",row.names = FALSE)
write.csv(FL_three,"FL_three.csv",row.names = F)
write.csv(NY_five,"NY_five.csv",row.names = FALSE)
write.csv(KY_one,"KY_one.csv",row.names = F)
Please share your code, thanx in advance!
CodePudding user response:
The 'out' list doesn't have any names because it was not named. If the names should come from the files part, we may name the output ('out') with the substring of file names
# returns all the file paths for csv
files <- list.files("C:\\mypath\\", pattern="\\.csv$", full.names = TRUE)
# get the substring of file names without the .csv part
filenms <- sub("\\.csv$", "", basename(files))
Now, we use the same code as in the OP's post or just loop over the files directly
out <- lapply(files, function(x) { # use lapply to loop over all files
this_data <- read.csv(x, header = TRUE)
aov_mod <- aov(revenue ~ dept, data = this_data)
tuk <- TukeyHSD(x= aov_mod)
# output of TukeyHSD is a list which can be summarised into tibble
# with tidy from broom
broom::tidy(tuk)
})
# set the names with filenms
names(out) <- filenms
If we want to write the output use imap/iwalk from purrr which is concise as .y returns the names and .x returns the value of the list
purrr::iwalk(out, ~ write.csv(.x, paste0(.y, ".csv"), row.names = FALSE))