I'd like to generate a PCA of my bulk RNAseq data, coloured by each of my variables in the DESeq2 object "vsd". My current code looks like this (to generate a single plot):
pcaData <- plotPCA(vsd, intgroup=c("Age", "BlastRate"), returnData=TRUE)
percentVar <- round(100 * attr(pcaData, "percentVar"))
ggplot(pcaData, aes(PC1, PC2, color=Age, shape=BlastRate))
geom_point(size=3)
xlab(paste0("PC1: ",percentVar[1],"% variance"))
ylab(paste0("PC2: ",percentVar[2],"% variance"))
geom_text(aes(label=name),hjust=-.2, vjust=0)
ggtitle("Principal Component Analysis")
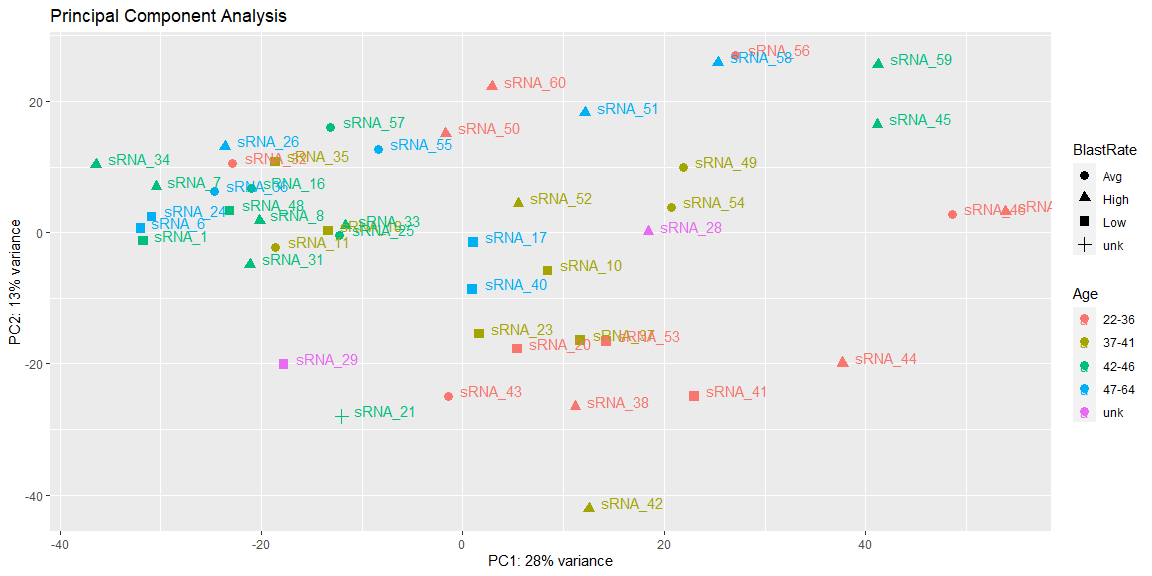 Can anyone suggest a method to loop through and swap "Age" with the other variable columns of vsd?
Can anyone suggest a method to loop through and swap "Age" with the other variable columns of vsd?
>head(colData(vsd),1)
DataFrame with 1 row and 14 columns
LibSize LibDiversity PercMapped Age SpermStatus SpConc SpMot Subject.Group PairedSample FertRate
<factor> <factor> <factor> <factor> <character> <factor> <factor> <factor> <factor> <factor>
sRNA_1 Low High High 42-46 unk unk unk Male-Male 2 Low
BlastRate RNABatch LibPrepBatch sizeFactor
<factor> <factor> <factor> <numeric>
sRNA_1 Low 1 6 0.929408
CodePudding user response:
Let's mock up some data, since the example you provide is too short and ill-formatted to do anything with. I'm assuming you have data of roughly the following structure:
library(ggplot2)
library(DESeq2)
dds <- makeExampleDESeqDataSet()
colData(dds) <- cbind(colData(dds), age = runif(ncol(dds), max = 50))
dds <- DESeq(dds)
vsd <- vst(dds, nsub = 200) # 200 is for example purposes
pcaData <- plotPCA(vsd, returnData = TRUE)
Next, we can select the column-names that you want to illustrate as a vector of strings and loop through them. You can use the .data pronoun to subset a particular column when using tidyverse-style non-standard evaluation.
vars <- tail(colnames(pcaData), 2)
plot_list <- lapply(vars, function(myvar) {
ggplot(pcaData, aes(PC1, PC2, colour = .data[[myvar]]))
geom_point()
})
# Just to show that there are multiple plots
patchwork::wrap_plots(plot_list)
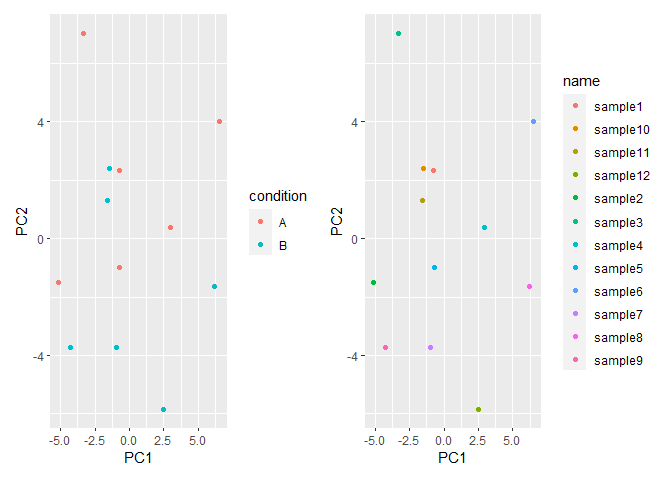
Created on 2022-03-09 by the reprex package (v2.0.1)
