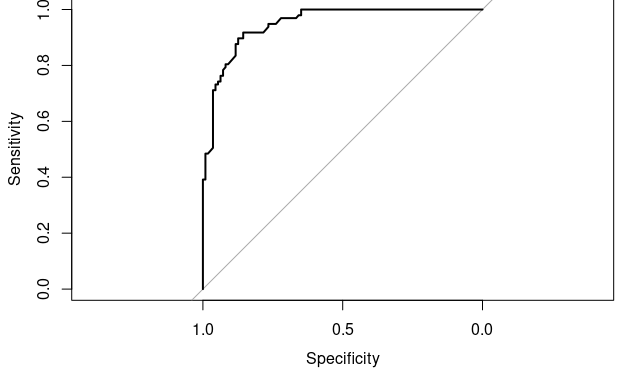
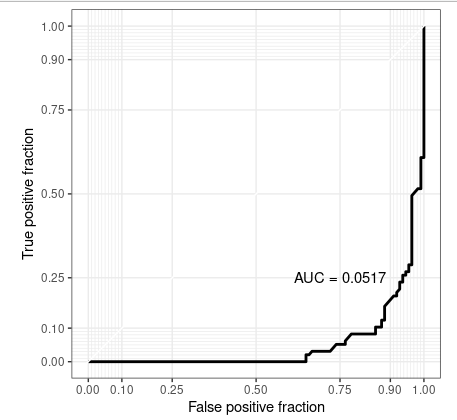
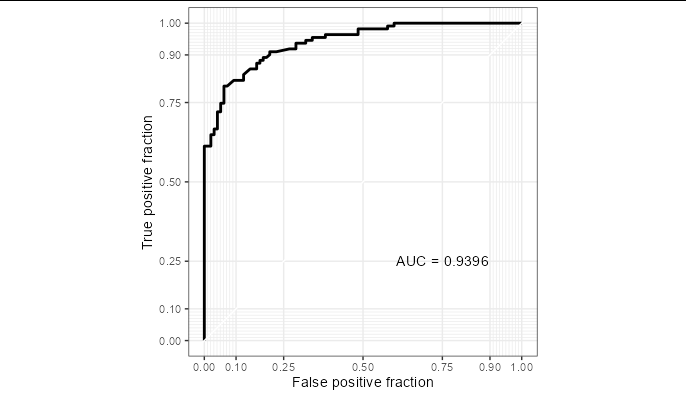
I supposed I have a training sent here.
library(caret)
library(mlbench)
library(plotROC)
library(pROC)
data(Sonar)
ctrl <- trainControl(method="cv",
summaryFunction=twoClassSummary,
classProbs=T,
savePredictions = T)
rfFit <- train(Class ~ ., data=Sonar,
method="rf", preProc=c("center", "scale"),
trControl=ctrl)
# Select a parameter setting
selectedIndices <- rfFit$pred$mtry == 2
and I would like to plot the ROC.
plot.roc(rfFit$pred$obs[selectedIndices],
rfFit$pred$M[selectedIndices])
however when I tried a ggplot2 approach it gives me something completely different.
g <- ggplot(rfFit$pred[selectedIndices, ], aes(m=M, d=factor(obs, levels = c("R", "M"))))
geom_roc(n.cuts=0)
coord_equal()
style_roc()
g annotate("text", x=0.75, y=0.25, label=paste("AUC =", round((calc_auc(g))$AUC, 4)))
Im doing something really wrong here but I can't figure out what it is. thanks.
CodePudding user response:
The order of your factor levels is ignored by geom_roc. Notice that whichever way round your assign your levels = c('R', 'M'), you get the warning:
#> Warning message:
#> In verify_d(data$d) : D not labeled 0/1, assuming M = 0 and R = 1!
This means you are getting the ROC of an 'anti-prediction' (i.e. the opposite of the prediction your model actually makes). Hence it is a mirror image of the actual ROC.
You need to explicitly convert the predictions to a numeric column of 1s and 0s:
g <- ggplot(rfFit$pred[selectedIndices, ],
aes(m=M, d= as.numeric(factor(obs, levels = c("R", "M"))) - 1))
geom_roc(n.cuts=0)
coord_equal()
style_roc()
g annotate("text", x=0.75, y=0.25,
label=paste("AUC =", round((calc_auc(g))$AUC, 4)))