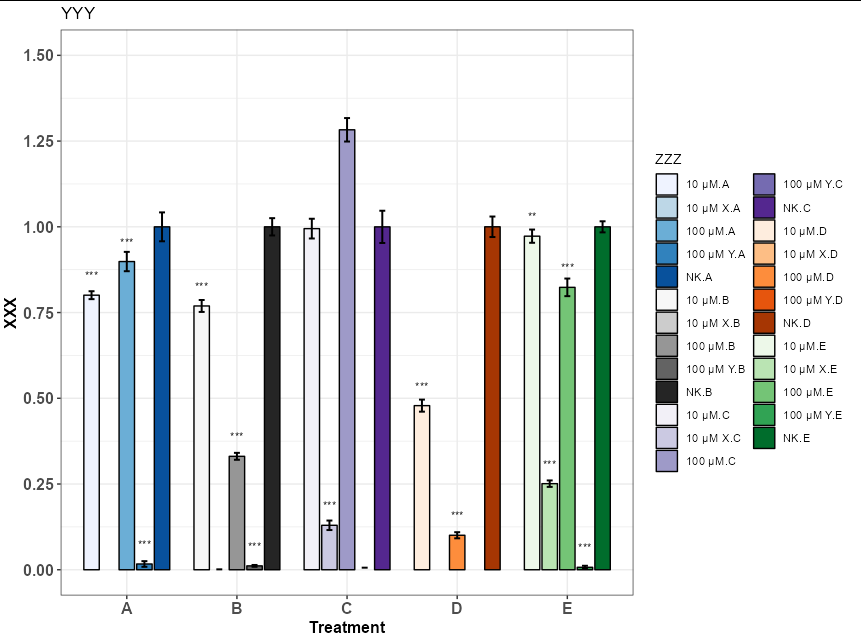
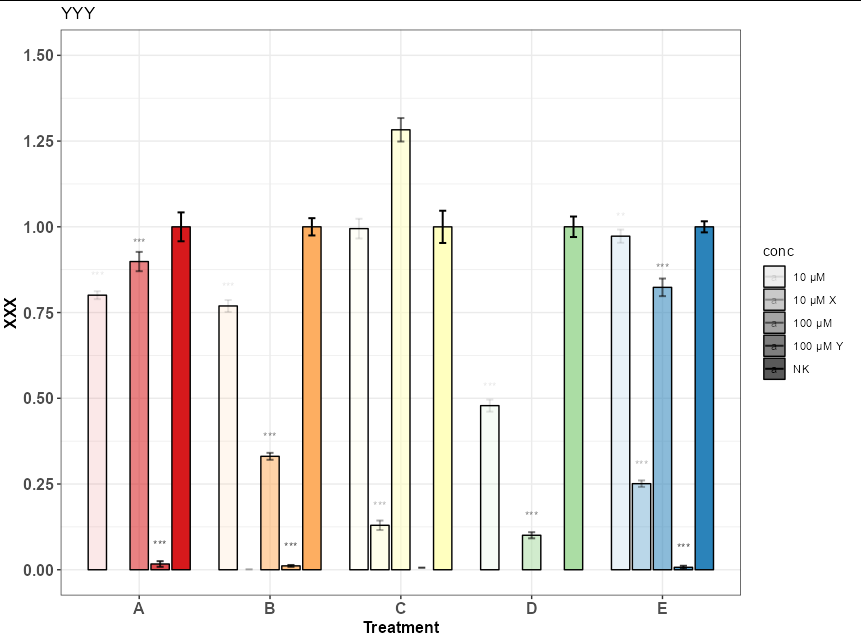
I am trying to put Several RColorBrewer Palettes that goes from lower tones to darker tones in one ggplot. But so far I've been unsuccessful and I've found that I can only use one.
My data set data:
data <- wrapr::build_frame(
"ID" , "Treatment", "conc" , "relabs" |
1 , "A" , "NK" , 0.9552 |
2 , "A" , "NK" , 1.016 |
3 , "A" , "NK" , 1.069 |
4 , "A" , "NK" , 1.029 |
5 , "A" , "NK" , 0.9992 |
6 , "A" , "NK" , 1.036 |
7 , "A" , "NK" , 0.9867 |
8 , "A" , "NK" , 0.9082 |
9 , "A" , "100 µM" , 0.9549 |
10 , "A" , "100 µM" , 0.9016 |
11 , "A" , "100 µM" , 0.9058 |
12 , "A" , "100 µM" , 0.9029 |
13 , "A" , "100 µM" , 0.8595 |
14 , "A" , "100 µM" , 0.8643 |
15 , "A" , "100 µM" , 0.8687 |
16 , "A" , "100 µM" , 0.9319 |
17 , "A" , "10 µM" , 0.8128 |
18 , "A" , "10 µM" , 0.805 |
19 , "A" , "10 µM" , 0.7765 |
20 , "A" , "10 µM" , 0.8065 |
21 , "A" , "10 µM" , 0.8153 |
22 , "A" , "10 µM" , 0.8045 |
23 , "A" , "10 µM" , 0.7827 |
24 , "A" , "10 µM" , 0.8017 |
25 , "A" , "10 µM X" , 0.00229 |
26 , "A" , "10 µM X" , 0.0002057 |
27 , "A" , "10 µM X" , -0.01033 |
28 , "A" , "10 µM X" , -0.003444 |
29 , "A" , "10 µM X" , -0.01401 |
30 , "A" , "10 µM X" , -0.007581 |
31 , "A" , "10 µM X" , -0.01063 |
32 , "A" , "10 µM X" , -0.01012 |
33 , "A" , "100 µM Y", 0.005991 |
34 , "A" , "100 µM Y", 0.01108 |
35 , "A" , "100 µM Y", 0.003925 |
36 , "A" , "100 µM Y", 0.02162 |
37 , "A" , "100 µM Y", 0.02916 |
38 , "A" , "100 µM Y", 0.01679 |
39 , "A" , "100 µM Y", 0.03044 |
40 , "A" , "100 µM Y", 0.01541 |
41 , "B" , "NK" , 1.038 |
42 , "B" , "NK" , 0.9651 |
43 , "B" , "NK" , 0.9948 |
44 , "B" , "NK" , 0.9688 |
45 , "B" , "NK" , 0.9727 |
46 , "B" , "NK" , 0.9985 |
47 , "B" , "NK" , 1.035 |
48 , "B" , "NK" , 1.027 |
49 , "B" , "100 µM" , 0.3466 |
50 , "B" , "100 µM" , 0.3429 |
51 , "B" , "100 µM" , 0.3131 |
52 , "B" , "100 µM" , 0.3302 |
53 , "B" , "100 µM" , 0.3204 |
54 , "B" , "100 µM" , 0.3265 |
55 , "B" , "100 µM" , 0.3238 |
56 , "B" , "100 µM" , 0.3425 |
57 , "B" , "10 µM" , 0.7703 |
58 , "B" , "10 µM" , 0.7484 |
59 , "B" , "10 µM" , 0.76 |
60 , "B" , "10 µM" , 0.7915 |
61 , "B" , "10 µM" , 0.7664 |
62 , "B" , "10 µM" , 0.7407 |
63 , "B" , "10 µM" , 0.7726 |
64 , "B" , "10 µM" , 0.8036 |
65 , "B" , "10 µM X" , -0.003965 |
66 , "B" , "10 µM X" , -0.001291 |
67 , "B" , "10 µM X" , 0.002101 |
68 , "B" , "10 µM X" , -0.001548 |
69 , "B" , "10 µM X" , 0.004782 |
70 , "B" , "10 µM X" , -0.006738 |
71 , "B" , "10 µM X" , -0.008429 |
72 , "B" , "10 µM X" , -0.009955 |
73 , "B" , "100 µM Y", 0.01063 |
74 , "B" , "100 µM Y", 0.008139 |
75 , "B" , "100 µM Y", 0.01149 |
76 , "B" , "100 µM Y", 0.01182 |
77 , "B" , "100 µM Y", 0.01418 |
78 , "B" , "100 µM Y", 0.009189 |
79 , "B" , "100 µM Y", 0.007849 |
80 , "B" , "100 µM Y", 0.0171 |
81 , "C" , "NK" , 0.9342 |
82 , "C" , "NK" , 1.033 |
83 , "C" , "NK" , 0.9425 |
84 , "C" , "NK" , 1 |
85 , "C" , "NK" , 1.082 |
86 , "C" , "NK" , 0.9697 |
87 , "C" , "NK" , 1.069 |
88 , "C" , "NK" , 0.9684 |
89 , "C" , "100 µM" , 1.31 |
90 , "C" , "100 µM" , 1.25 |
91 , "C" , "100 µM" , 1.305 |
92 , "C" , "100 µM" , 1.28 |
93 , "C" , "100 µM" , 1.293 |
94 , "C" , "100 µM" , 1.256 |
95 , "C" , "100 µM" , 1.35 |
96 , "C" , "100 µM" , 1.219 |
97 , "C" , "10 µM" , 0.9741 |
98 , "C" , "10 µM" , 1.066 |
99 , "C" , "10 µM" , 0.9849 |
100 , "C" , "10 µM" , 0.9737 |
101 , "C" , "10 µM" , 0.9619 |
102 , "C" , "10 µM" , 0.989 |
103 , "C" , "10 µM" , 0.9821 |
104 , "C" , "10 µM" , 1.026 |
105 , "C" , "10 µM X" , 0.137 |
106 , "C" , "10 µM X" , 0.1283 |
107 , "C" , "10 µM X" , 0.09757 |
108 , "C" , "10 µM X" , 0.1522 |
109 , "C" , "10 µM X" , 0.1411 |
110 , "C" , "10 µM X" , 0.1377 |
111 , "C" , "10 µM X" , 0.1222 |
112 , "C" , "10 µM X" , 0.1209 |
113 , "C" , "100 µM Y", -0.00434 |
114 , "C" , "100 µM Y", -0.009208 |
115 , "C" , "100 µM Y", 0.01106 |
116 , "C" , "100 µM Y", -0.0005099 |
117 , "C" , "100 µM Y", 0.001142 |
118 , "C" , "100 µM Y", -0.002433 |
119 , "C" , "100 µM Y", 0.009931 |
120 , "C" , "100 µM Y", -0.01025 |
121 , "D" , "NK" , 1.046 |
122 , "D" , "NK" , 1.032 |
123 , "D" , "NK" , 0.9685 |
124 , "D" , "NK" , 0.9981 |
125 , "D" , "NK" , 1.005 |
126 , "D" , "NK" , 1.001 |
127 , "D" , "NK" , 0.9329 |
128 , "D" , "NK" , 1.017 |
129 , "D" , "100 µM" , 0.1012 |
130 , "D" , "100 µM" , 0.1177 |
131 , "D" , "100 µM" , 0.09581 |
132 , "D" , "100 µM" , 0.09372 |
133 , "D" , "100 µM" , 0.1143 |
134 , "D" , "100 µM" , 0.1019 |
135 , "D" , "100 µM" , 0.08676 |
136 , "D" , "100 µM" , 0.09314 |
137 , "D" , "10 µM" , 0.461 |
138 , "D" , "10 µM" , 0.4717 |
139 , "D" , "10 µM" , 0.4536 |
140 , "D" , "10 µM" , 0.487 |
141 , "D" , "10 µM" , 0.5137 |
142 , "D" , "10 µM" , 0.4936 |
143 , "D" , "10 µM" , 0.4574 |
144 , "D" , "10 µM" , 0.4904 |
145 , "D" , "10 µM X" , -0.02192 |
146 , "D" , "10 µM X" , -0.02502 |
147 , "D" , "10 µM X" , -0.0238 |
148 , "D" , "10 µM X" , -0.01711 |
149 , "D" , "10 µM X" , -0.02345 |
150 , "D" , "10 µM X" , -0.01186 |
151 , "D" , "10 µM X" , -0.004447 |
152 , "D" , "10 µM X" , -0.01209 |
153 , "D" , "100 µM Y", -0.01495 |
154 , "D" , "100 µM Y", -0.01741 |
155 , "D" , "100 µM Y", -0.0101 |
156 , "D" , "100 µM Y", -0.007783 |
157 , "D" , "100 µM Y", 0.004533 |
158 , "D" , "100 µM Y", -0.01373 |
159 , "D" , "100 µM Y", -0.02207 |
160 , "D" , "100 µM Y", -0.01263 |
161 , "E" , "NK" , 1.03 |
162 , "E" , "NK" , 0.9683 |
163 , "E" , "NK" , 0.9915 |
164 , "E" , "NK" , 0.9887 |
165 , "E" , "NK" , 1.019 |
166 , "E" , "NK" , 1.007 |
167 , "E" , "NK" , 0.9909 |
168 , "E" , "NK" , 1.004 |
169 , "E" , "100 µM" , 0.7583 |
170 , "E" , "100 µM" , 0.8541 |
171 , "E" , "100 µM" , 0.822 |
172 , "E" , "100 µM" , 0.8506 |
173 , "E" , "100 µM" , 0.8122 |
174 , "E" , "100 µM" , 0.8442 |
175 , "E" , "100 µM" , 0.831 |
176 , "E" , "100 µM" , 0.8153 |
177 , "E" , "10 µM" , 0.9815 |
178 , "E" , "10 µM" , 0.9623 |
179 , "E" , "10 µM" , 0.97 |
180 , "E" , "10 µM" , 0.9798 |
181 , "E" , "10 µM" , 0.967 |
182 , "E" , "10 µM" , 0.9825 |
183 , "E" , "10 µM" , 1.01 |
184 , "E" , "10 µM" , 0.9284 |
185 , "E" , "10 µM X" , 0.2576 |
186 , "E" , "10 µM X" , 0.2454 |
187 , "E" , "10 µM X" , 0.2467 |
188 , "E" , "10 µM X" , 0.2544 |
189 , "E" , "100 µM Y", 0.005576 |
190 , "E" , "100 µM Y", 0.01025 |
191 , "E" , "100 µM Y", 0.00863 |
192 , "E" , "100 µM Y", 0.004152 )
data_summary <-
data %>%
group_by(Treatment, conc) %>%
dplyr::summarize(relabs_avg = mean(relabs),
relabs_sd = sd(relabs),
relabs_median = median(relabs),
relabs_mad = mad(relabs),
relabs_q1 = quantile(relabs, probs = c(0.25)),
relabs_q3 = quantile(relabs, probs = c(0.75)),
size = n()) %>%
dplyr::mutate(across(where(is.numeric), ~round(., digits = 3)))
data_summary
alpha <- 0.05
data_full <-
data %>%
group_by(Treatment, conc) %>%
dplyr:: summarize(mean = mean(relabs),
median = median(relabs),
lower = mean(relabs) - qt(1- alpha/2, (n() - 1))*sd(relabs)/sqrt(n()),
upper = mean(relabs) qt(1- alpha/2, (n() - 1))*sd(relabs)/sqrt(n()))
data_full
df<- merge(data_summary, data_full)
df
df_t_test <-
df_full %>%
group_by(Treatment, conc) %>%
do(tidy(t.test(.$relabs,
mu = 1 ,
alt = "less",
conf.level = 0.95, var.equal = FALSE)))
df_t_test
df_full<- merge(data, df)
df_full
df_full<- merge(data_full, df_t_test)
df_full
What I'm currently using:
df_full$Label <- NA
df_full$Label[df_full$mean <0]<-'ND'
df_full$Label[df_full$p.value<0.001 & is.na(df_full$Label)]<-'***'
df_full$Label[df_full$p.value<0.01 & is.na(df_full$Label)]<-'**'
df_full$Label[df_full$p.value<0.05 & is.na(df_full$Label)]<-'*'
breaks_y =c(0, 0.25, 0.5, 0.75, 1, 1.25, 1.5)
df_full$Label <- NA
df_full$Label[df_full$mean <0]<-'ND'
df_full$Label[df_full$p.value<0.001 & is.na(df_full$Label)]<-'***'
df_full$Label[df_full$p.value<0.01 & is.na(df_full$Label)]<-'**'
df_full$Label[df_full$p.value<0.05 & is.na(df_full$Label)]<-'*'
plot <-
ggplot(df_full, aes(x = factor (Treatment, level = c("A","B", "C", "D", "E")), y = mean, fill = conc))
geom_col(color = "black", position = position_dodge(0.8), width = 0.7)
geom_errorbar(aes(ymax = upper, ymin = lower), width = 0.27, position = position_dodge(0.8), color = "black", size = 0.7)
geom_text(aes(label = Label, group = conc),size = 3, position = position_dodge(width =0.8), color = "black", vjust =-2)
labs(x = "Treatment", y = "XXX", title = "YYY ", color = "ZZZ", fill = "ZZZ")
scale_y_continuous(limits = c(0, 1.5), breaks = breaks_y)
theme_bw()
theme(axis.text = element_text(size = 12, face = "bold"),
axis.title.y = element_text(size = 12, face ="bold"),
axis.title.x = element_text(size = 12, face ="bold"))
plot scale_fill_brewer(palette = "Blues")
Is there a way to put color palette "Blues" on A Treatment, "Greys" on B Treatment and so on? Or some kind of manual way to do that I wasn't able to find?
CodePudding user response:
You would really have to create your own combined Brewer palette and apply it to the interaction of the two grouping variables (conc and Treatment)
fills <- c(sapply(c("Blues", "Greys", "Purples", "Oranges", "Greens"),
function(x) brewer.pal(5, x)))
ggplot(df_full, aes(x = factor (Treatment, level = c("A","B", "C", "D", "E")),
y = mean, fill = interaction(conc, Treatment)))
geom_col(color = "black", position = position_dodge(0.8), width = 0.7)
geom_errorbar(aes(ymax = upper, ymin = lower), width = 0.27,
position = position_dodge(0.8), color = "black", size = 0.7)
geom_text(aes(label = Label, group = conc),size = 3,
position = position_dodge(width =0.8), color = "black", vjust =-2)
labs(x = "Treatment", y = "XXX", title = "YYY ", color = "ZZZ", fill = "ZZZ")
scale_y_continuous(limits = c(0, 1.5), breaks = breaks_y)
theme_bw()
theme(axis.text = element_text(size = 12, face = "bold"),
axis.title.y = element_text(size = 12, face ="bold"),
axis.title.x = element_text(size = 12, face ="bold"))
scale_fill_manual(values = fills)
Of course, the problem here is that your legend is now quite unwieldy. However, for a discrete color scale, it's difficult to get round this.
Possibly the cleanest way to achieve a similar effect is to fill according to treatment and use the alpha scale for conc
ggplot(df_full, aes(x = factor (Treatment, level = c("A","B", "C", "D", "E")),
y = mean, fill = Treatment, alpha = conc))
geom_col(color = "black", position = position_dodge(0.8), width = 0.7)
geom_errorbar(aes(ymax = upper, ymin = lower), width = 0.27,
position = position_dodge(0.8), color = "black", size = 0.7)
geom_text(aes(label = Label, group = conc),size = 3,
position = position_dodge(width =0.8), color = "black", vjust =-2)
labs(x = "Treatment", y = "XXX", title = "YYY ", color = "ZZZ", fill = "ZZZ")
scale_y_continuous(limits = c(0, 1.5), breaks = breaks_y)
theme_bw()
theme(axis.text = element_text(size = 12, face = "bold"),
axis.title.y = element_text(size = 12, face ="bold"),
axis.title.x = element_text(size = 12, face ="bold"))
scale_fill_brewer(palette ="Spectral" )
guides(fill = guide_none())