I have data.table as follows and I want to smooth the z-axis so that it creates a smooth surface when plotted on the x and y-axis.
library(data.table)
temp_dt = structure(list(x = c(160, 175, 185, 200, 220, 240, 250, 260,
270, 280, 285, 290, 290, 295, 300, 300, 300, 305, 305, 310, 310,
310, 315, 315, 315, 320, 320, 320, 325, 325, 325, 330, 330, 330,
335, 335, 340, 340, 340, 345, 345, 350, 350, 350, 350, 355, 355,
355, 360, 360, 360, 360, 365, 365, 365, 365, 370, 370, 370, 375,
375, 375, 380, 380, 380, 380, 385, 385, 385, 385, 385, 390, 390,
390, 390, 390, 390, 395, 395, 395, 395, 395, 400, 400, 400, 400,
400, 405, 405, 405, 405, 405, 410, 410, 410, 410, 410, 410, 410,
415, 415, 415, 415, 415, 420, 420, 420, 420, 420, 420, 420, 425,
425, 425, 425, 425, 425, 430, 430, 430, 430, 430, 430, 430, 430,
432.5, 435, 435, 435, 435, 435, 435, 435, 437.5, 437.5, 437.5,
440, 440, 440, 440, 440, 440, 440, 440, 440, 442.5, 442.5, 445,
445, 445, 445, 445, 445, 445, 447.5, 447.5, 447.5, 450, 450,
450, 450, 450, 450, 450, 450, 452.5, 452.5, 452.5, 455, 455,
455, 455, 455, 455, 457.5, 460, 460, 460, 460, 460, 460, 460,
460, 462.5, 465, 465, 465, 465, 465, 465, 465, 465, 470, 470,
470, 470, 470, 470, 470, 470, 475, 475, 475, 475, 475, 475, 475,
480, 480, 480, 480, 480, 480, 480, 480, 485, 485, 485, 485, 485,
490, 490, 490, 490, 490, 490, 490, 490, 495, 495, 495, 495, 495,
500, 500, 500, 500, 500, 500, 500, 500, 500, 505, 505, 505, 505,
505, 510, 510, 510, 510, 510, 515, 515, 515, 515, 520, 520, 520,
520, 520, 520, 520, 525, 525, 525, 530, 530, 535, 535, 540, 540,
540, 545, 550, 555, 560, 560, 560, 565, 570, 580, 580, 580, 590,
600, 600, 600, 600, 605, 620, 640, 640, 640, 660, 660, 680, 680,
720), y = c(0.8361, 0.8361, 0.8361, 0.8361, 0.8361, 0.8361, 0.8361,
0.8361, 0.8361, 0.8361, 0.1556, 0.1556, 0.8361, 0.8361, 0.1556,
0.4083, 0.8361, 0.1556, 0.8361, 0.1556, 0.4083, 0.8361, 0.1556,
0.4083, 0.8361, 0.1556, 0.4083, 0.8361, 0.1556, 0.4083, 0.8361,
0.1556, 0.4083, 0.8361, 0.1556, 0.8361, 0.1556, 0.4083, 0.8361,
0.1556, 0.8361, 0.0556, 0.1556, 0.4083, 0.8361, 0.1556, 0.4083,
0.8361, 0.0556, 0.1556, 0.4083, 0.8361, 0.0389, 0.1556, 0.4083,
0.8361, 0.0556, 0.1556, 0.8361, 0.1556, 0.4083, 0.8361, 0.0556,
0.1556, 0.4083, 0.8361, 0.0194, 0.0556, 0.1556, 0.4083, 0.8361,
0.0194, 0.0556, 0.0778, 0.1556, 0.4083, 0.8361, 0.0194, 0.0389,
0.0556, 0.1556, 0.4083, 0.0194, 0.0556, 0.1556, 0.4083, 0.8361,
0.0194, 0.0556, 0.0778, 0.1556, 0.4083, 0.0194, 0.0389, 0.0556,
0.0778, 0.1556, 0.4083, 0.8361, 0.0194, 0.0389, 0.0556, 0.1556,
0.4083, 0.0194, 0.0389, 0.0556, 0.0778, 0.1556, 0.4083, 0.8361,
0.0194, 0.0389, 0.0556, 0.0972, 0.1556, 0.4083, 0.0194, 0.0389,
0.0556, 0.0778, 0.0972, 0.1556, 0.4083, 0.8361, 0.0194, 0.0194,
0.0389, 0.0556, 0.0778, 0.0972, 0.1556, 0.4083, 0.0194, 0.0389,
0.0556, 0.0194, 0.0389, 0.0556, 0.0778, 0.0972, 0.1556, 0.4083,
0.6611, 0.8361, 0.0194, 0.0556, 0.0194, 0.0389, 0.0556, 0.0778,
0.0972, 0.1556, 0.4083, 0.0194, 0.0389, 0.0556, 0.0194, 0.0389,
0.0556, 0.0778, 0.0972, 0.1556, 0.4083, 0.8361, 0.0194, 0.0389,
0.0556, 0.0194, 0.0389, 0.0556, 0.0778, 0.1556, 0.4083, 0.0194,
0.0194, 0.0389, 0.0556, 0.0778, 0.0972, 0.1556, 0.4083, 0.8361,
0.0194, 0.0194, 0.0389, 0.0556, 0.0778, 0.0972, 0.1556, 0.4083,
0.6611, 0.0194, 0.0389, 0.0556, 0.0778, 0.0972, 0.1556, 0.4083,
0.8361, 0.0194, 0.0389, 0.0556, 0.0778, 0.0972, 0.1556, 0.4083,
0.0194, 0.0389, 0.0556, 0.0778, 0.0972, 0.1556, 0.4083, 0.8361,
0.0194, 0.0389, 0.0556, 0.0778, 0.1556, 0.0194, 0.0389, 0.0556,
0.0778, 0.1167, 0.1556, 0.4083, 0.8361, 0.0194, 0.0556, 0.0778,
0.0972, 0.1556, 0.0194, 0.0389, 0.0556, 0.0778, 0.1167, 0.1556,
0.4083, 0.6611, 0.8361, 0.0194, 0.0389, 0.0556, 0.0972, 0.1556,
0.0194, 0.0389, 0.0556, 0.0778, 0.1556, 0.0389, 0.0556, 0.0778,
0.1556, 0.0194, 0.0556, 0.1167, 0.1556, 0.4083, 0.6611, 0.8361,
0.0556, 0.0972, 0.1556, 0.0556, 0.1556, 0.0556, 0.1556, 0.1556,
0.4083, 0.8361, 0.1556, 0.1556, 0.1556, 0.1556, 0.4083, 0.8361,
0.1556, 0.1556, 0.1556, 0.4083, 0.8361, 0.1556, 0.0778, 0.1556,
0.4083, 0.8361, 0.1556, 0.8361, 0.1556, 0.4083, 0.8361, 0.4083,
0.8361, 0.4083, 0.8361, 0.8361), z = c(298.25, 283.5, 273.7,
258.8, 239.1, 219.5, 210.15, 200.75, 191.35, 181.75, 174, 169.1,
172.75, 167.95, 158.85, 160.5, 163.6, 154.45, 159.05, 149.2,
150.65, 154, 144.6, 145.85, 149.05, 139.55, 141.35, 144.9, 134.7,
136.4, 141.15, 129.65, 132.15, 136.85, 125, 133.25, 119.9, 122.9,
128.55, 114.95, 124.7, 108.4, 110.1, 113.1, 120.45, 105.4, 109.15,
115.7, 98.55, 101.05, 104.4, 112.25, 93.7, 95.65, 100.35, 108.05,
88.8, 90.7, 104.95, 86.25, 91.9, 100.75, 79, 81.4, 87.5, 96.85,
73.7, 73.2, 77.1, 83.6, 93.2, 68.35, 69.25, 69.15, 72.6, 79.5,
90.15, 63.75, 63.8, 63.19, 68.25, 75.6, 58.45, 58.8, 62.9, 70.4,
84.1, 53.19, 54.5, 54.8, 59.35, 68.3, 48.45, 49.25, 48.95, 50.75,
54.35, 64.55, 76.35, 43.7, 44.4, 45.2, 50.9, 61, 38.8, 39.6,
40.2, 41.2, 46.4, 56.45, 69.7, 33.4, 35.1, 36.1, 38.25, 42.9,
53.95, 28.9, 30.5, 31.65, 32.6, 34.2, 38.95, 50.55, 64.65, 26.8,
24.45, 25.95, 27.5, 28.75, 30.85, 35.75, 47.35, 21.95, 24.05,
25.7, 20.2, 22.2, 23.4, 25.25, 27.25, 32.55, 43.75, 52.35, 59.15,
17.75, 21.4, 15.9, 18.1, 20.35, 21.4, 24.65, 30.2, 41.3, 13.85,
16.95, 18.4, 12.2, 14.85, 16.9, 17.7, 21.1, 26.35, 38.25, 52.8,
10.4, 13.65, 15.5, 9.05, 12.15, 14.05, 15.8, 23.6, 35.75, 7.65,
6.4, 9.65, 11.55, 12.9, 16, 21.25, 33.15, 48, 5.8, 4.45, 7.4,
9.45, 10.9, 13.9, 18.85, 30.95, 38.9, 3.125, 5.5, 7.05, 9, 11.75,
16.1, 28.8, 43.35, 2.15, 4.25, 5.8, 7.25, 9.25, 14.5, 26.45,
1.55, 2.75, 4.6, 5.6, 8.3, 12.8, 24.5, 39.4, 1.05, 2.3, 3.6,
4.8, 11, 0.7, 2.15, 2.8, 4.2, 6.95, 9.65, 20.75, 35.25, 0.775,
2.3, 3.675, 5.55, 8.4, 0.25, 1.175, 1.8, 3.1, 5.25, 7, 17.65,
24.8, 32.4, 0.425, 1.05, 1.45, 3.45, 6.15, 1.125, 0.975, 1.225,
2.45, 5.55, 0.6, 1.025, 2.15, 4.6, 0.725, 0.725, 3.025, 4.05,
12.55, 19.2, 25.65, 1.2, 2.125, 4.05, 1.25, 3.075, 1.125, 2.675,
2.275, 7.9, 19.7, 1.8, 1.825, 1.575, 1.475, 6, 15.65, 1.3, 1.775,
1.475, 4.3, 12.2, 0.95, 0.75, 0.65, 3.5, 10.1, 0.875, 7.1, 0.85,
1.85, 5.95, 2.1, 4.45, 0.95, 3.475, 2.375)), row.names = c(NA,
-299L), class = c("data.table", "data.frame"))
Can someone help me to -
- Transform/ smooth the surface (z-axis) non-linearly. I have tried the following but it returns NA values for the z-axis.
library(akima)
with(temp_dt, akima::interp(x=x, y=y, z=z, nx = length(unique(x)), ny = length(unique(y)), duplicate = "mean", linear=FALSE)
- Draw the 3D view of the smoothed surface. I have tried
perspfunction but it throws an error -increasing 'x' and 'y' values expected.
with(temp_dt, persp(x=x, y=y, z=z))
CodePudding user response:
You can do:
library(interp)
mat <- interp(temp_dt$x, temp_dt$y, temp_dt$z)
persp(mat$x, mat$y, mat$z)
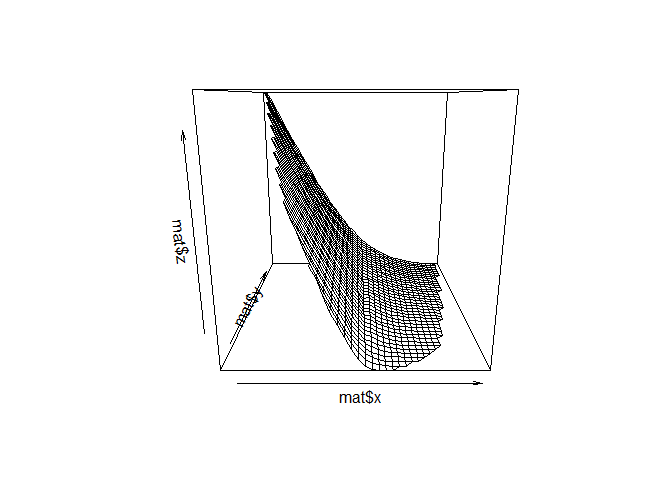
The reason why there are NA values in the final matrix is that the z values will not extrapolate outside the convex hull of your points.
Note that instead of using persp, you can have a fully rotatable 3D plot if you use plotly
library(plotly)
fig <- plot_ly(x = mat$x, y = mat$y, z = ~mat$z)
add_surface(fig)
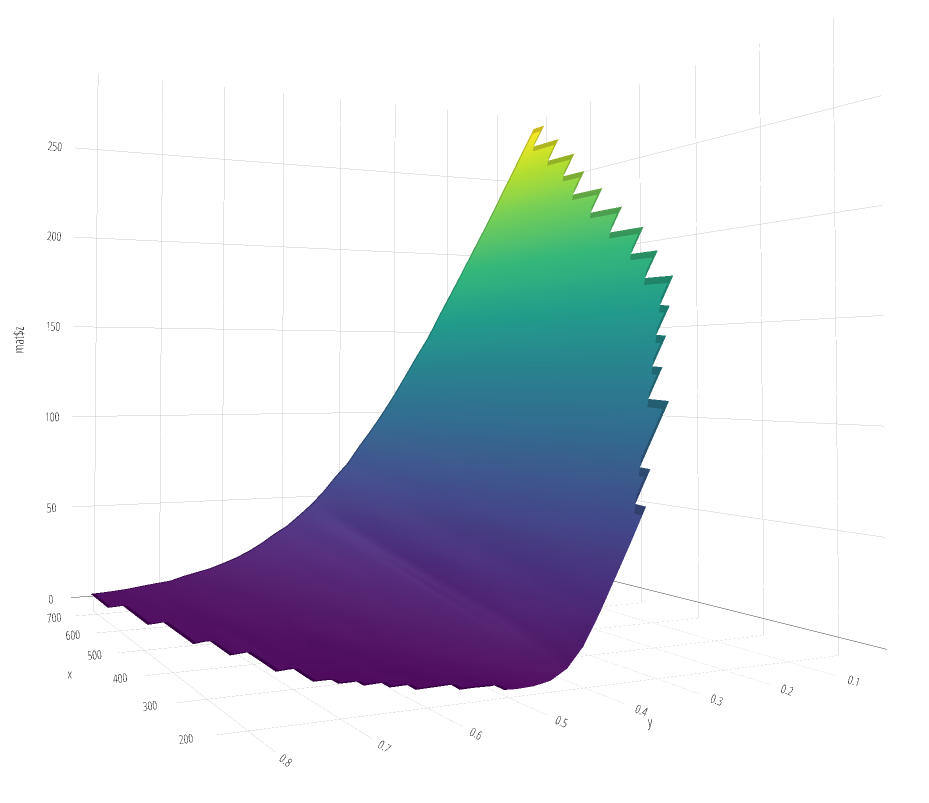 Created on 2022-03-26 by the reprex package (v2.0.1)
Created on 2022-03-26 by the reprex package (v2.0.1)
