I want to interpolate a 3D array along the first dimension.
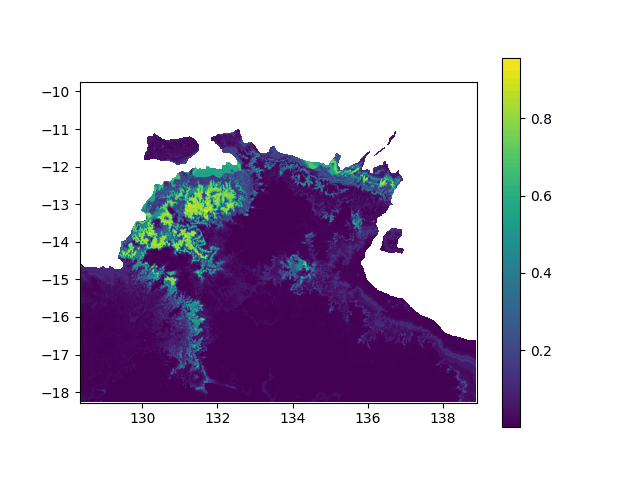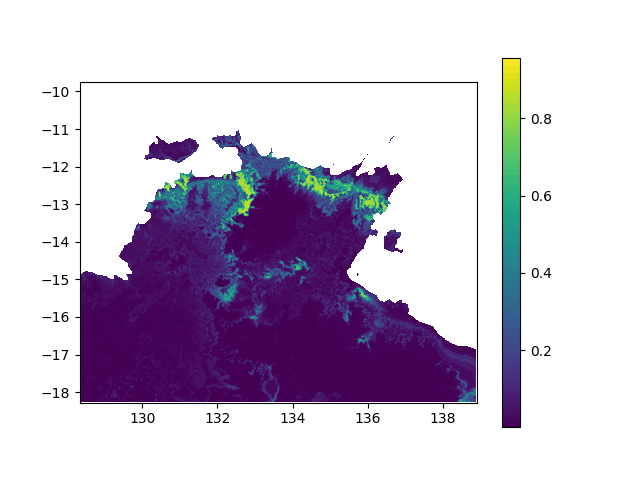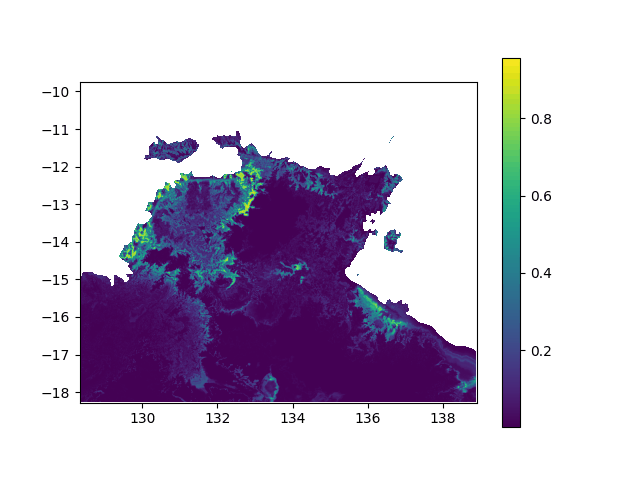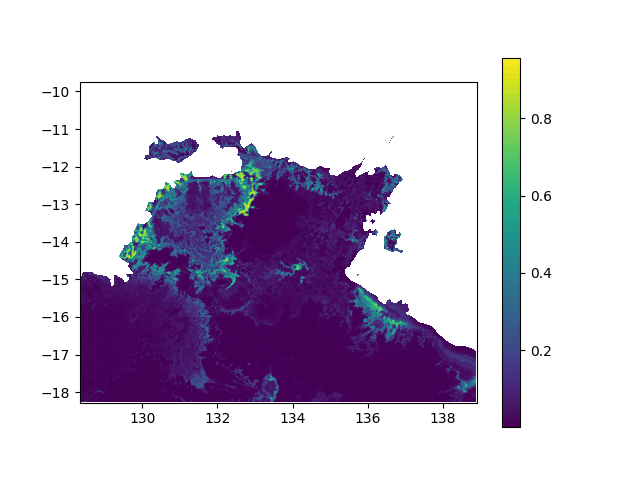
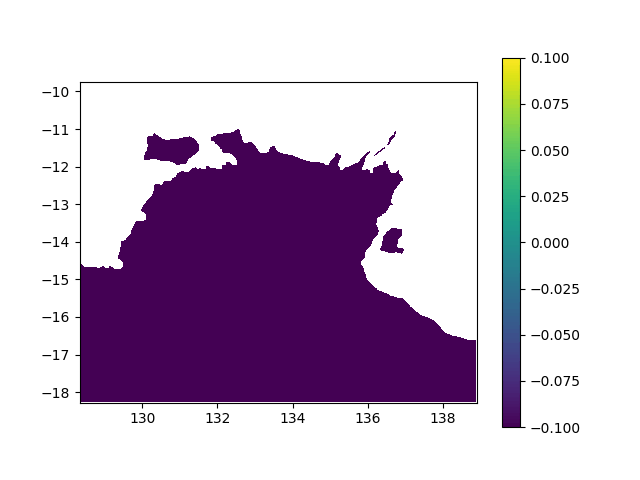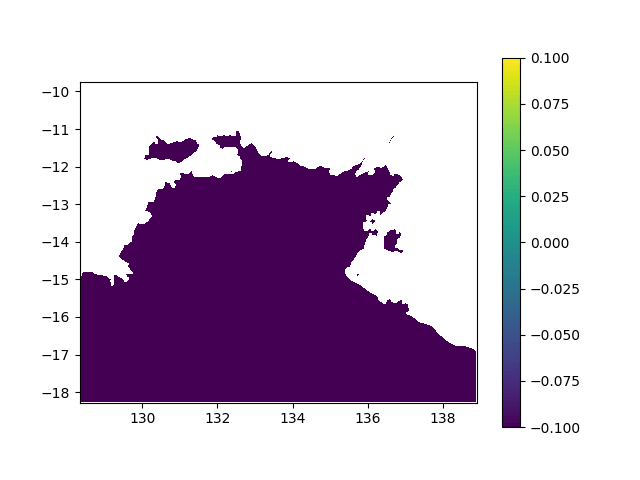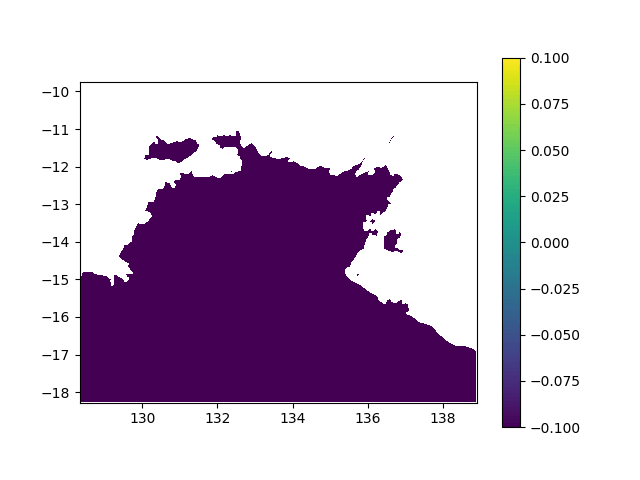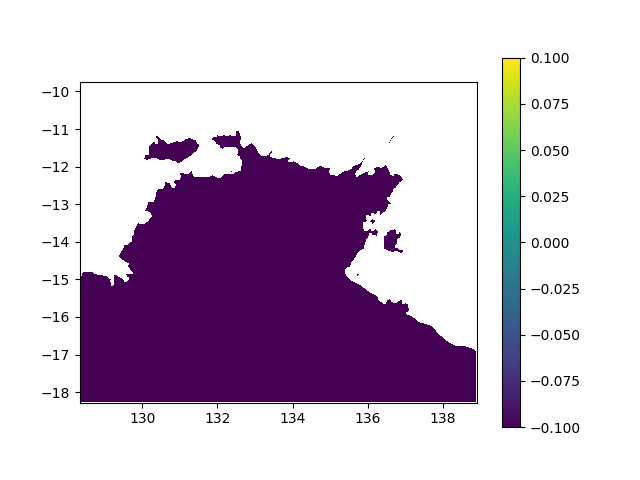
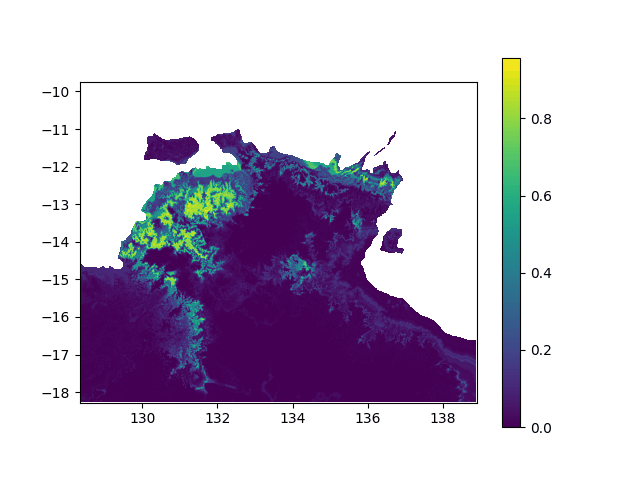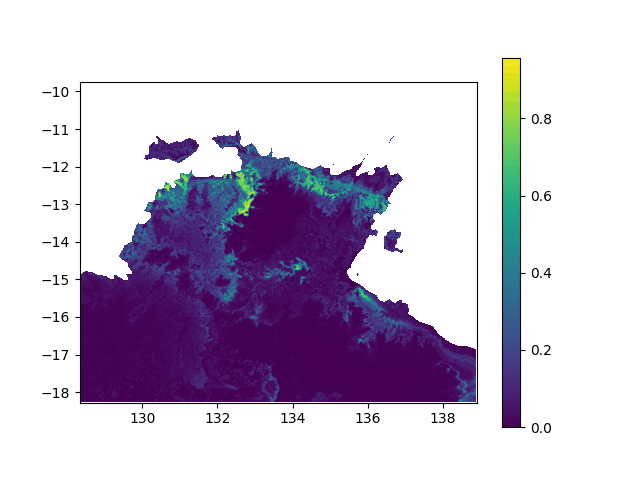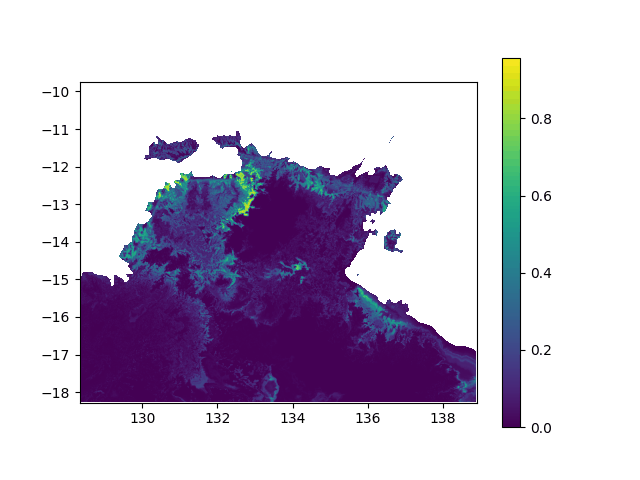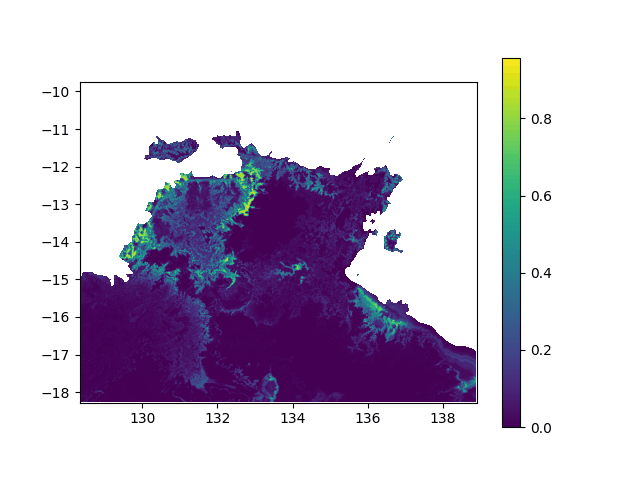
In terms of data, it means I want to interpolated missing times in a geographic value, in other terms smoothing a bit this animation:
I do this by calling:
new = ma.apply_along_axis(func1d=masked_interpolation, axis=0, arr=dst_data, x=missing_bands, xp=known_bands)
Where the interpolation function is the following:
def masked_interpolation(data, x, xp, propagate_mask=True):
import math
import numpy as np
import numpy.ma as ma
# The x-coordinates (missing times) at which to evaluate the interpolated values.
assert len(x) >= 1
# The x-coordinates (existing times) of the data points (where returns a tuple because each element of the tuple refers to a dimension.)
assert len(xp) >= 2
# The y-coordinates (value at existing times) of the data points, that is the valid entries
fp = np.take(data, xp)
assert len(fp) >= 2
# Returns the one-dimensional piecewise linear interpolant to a function with given discrete data points (xp, fp), evaluated at x.
new_y = np.interp(x, xp, fp.filled(np.nan))
# interpolate mask & apply to interpolated data
if propagate_mask:
new_mask = data.mask[:]
new_mask[new_mask] = 1
new_mask[~new_mask] = 0
# the mask y values at existing times
new_fp = np.take(new_mask, xp)
new_mask = np.interp(x, xp, new_fp)
new_y = np.ma.masked_array(new_y, new_mask > 0.5)
print(new_y) # ----> that seems legit
data[x] = new_y # ----> from here it goes wrong
return data
When printing new_y, the interpolated values seem consistent (spread across [0,1] interval, what I want). However, when I print the final output (the new array), it's definitely smoother (more bands) but all the non-masked values are changed to -0.1 (what does not make any sense):
The code to write that to a raster file is:
# Writing the new raster
meta = source.meta
meta.update({'count' : dst_shape[0] })
meta.update({'nodata' : source.nodata})
meta.update(fill_value = source.nodata)
assert new.shape == (meta['count'],meta['height'],meta['width'])
with rasterio.open(outputFile, "w", **meta) as dst:
dst.write(new.filled(fill_value=source.nodata))
CodePudding user response:
It was quite tricky to figure out. What happens is that the interpolation function has to fill with nans so the interpolation works, but then replace remaining nans (coming eg from when the whole fp vector is nan) with finite values. Then applying the interpolated mask will hide these values anyway. Here is how it goes:
def masked_interpolation(data, x, xp, propagate_mask=True):
import math
import numpy as np
import numpy.ma as ma
# The x-coordinates (missing times) at which to evaluate the interpolated values.
assert len(x) >= 1
# The x-coordinates (existing times) of the data points (where returns a tuple because each element of the tuple refers to a dimension.)
assert len(xp) >= 2
# The y-coordinates (value at existing times) of the data points, that is the valid entries
fp = np.take(data, xp)
assert len(fp) >= 2
# Returns the one-dimensional piecewise linear interpolant to a function with given discrete data points (xp, fp), evaluated at x.
new_y = np.interp(x, xp, fp.filled(np.nan))
np.nan_to_num(new_y, copy=False)
# interpolate mask & apply to interpolated data
if propagate_mask:
new_mask = data.mask[:]
new_mask[new_mask] = 1
new_mask[~new_mask] = 0
# the mask y values at existing times
new_fp = np.take(new_mask, xp)
new_mask = np.interp(x, xp, new_fp)
new_y = np.ma.masked_array(new_y, new_mask > 0.5)
data[x] = new_y
return data