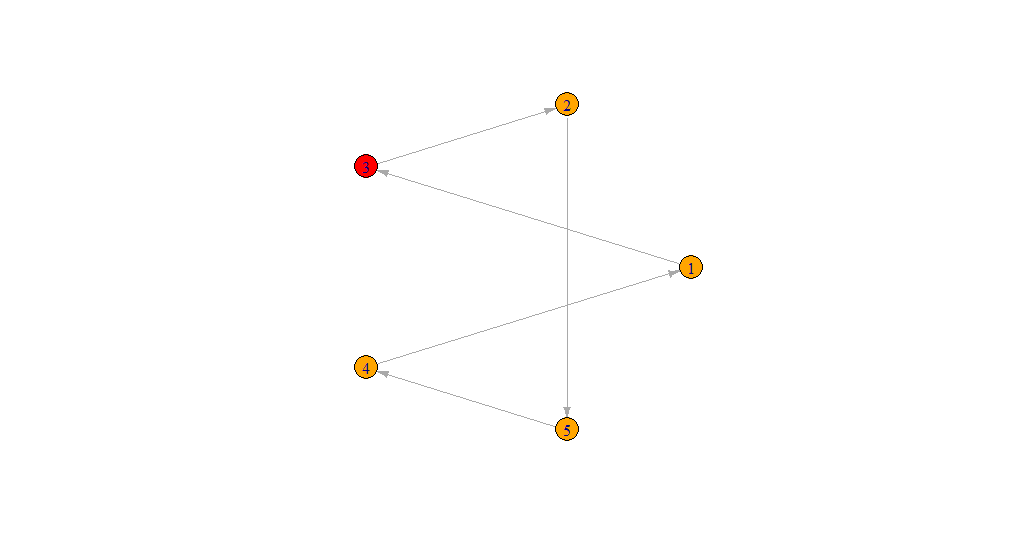
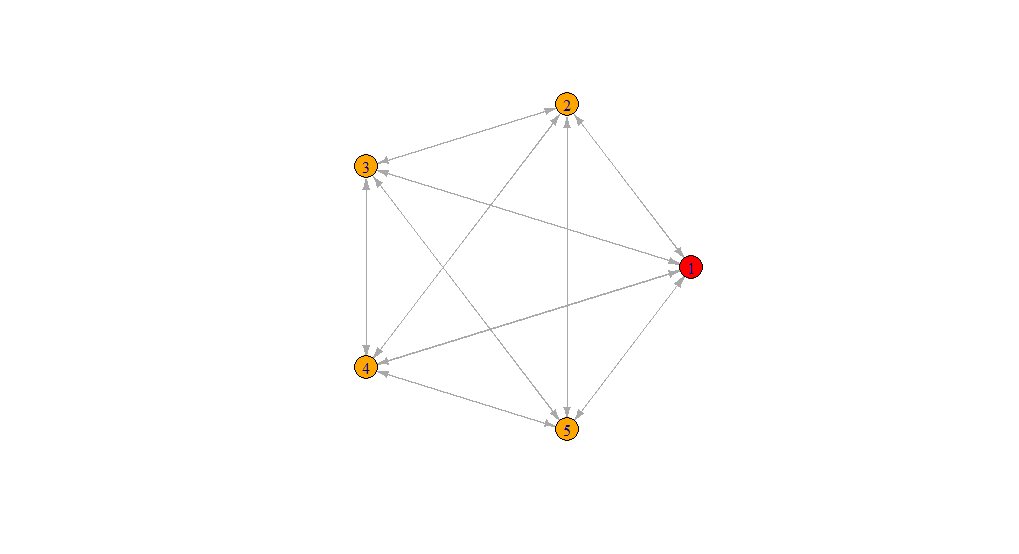
I have the following network graph:
library(tidyverse)
library(igraph)
set.seed(123)
n=5
data = tibble(d = paste(1:n))
relations = data.frame(tibble(
from = sample(data$d),
to = lead(from, default=from[1]),
))
graph = graph_from_data_frame(relations, directed=T, vertices = data)
V(graph)$color <- ifelse(data$d == relations$from[1], "red", "orange")
plot(graph, layout=layout.circle, edge.arrow.size = 0.2)
I want to connect each Node to every Node on this graph - I can do this manually by redefining the "relations" data frame:
relations_1 = data.frame("from" = c(1,1,1,1,2,2,2,2,3,3,3,3,4,4,4,4,5,5,5,5), "to" = c(2,3,4,5,1,3,4,5,1,2,4,5,1,2,3,5,1,2,3,4))
Then, I can re-run the network graph:
graph = graph_from_data_frame(relations_1, directed=T, vertices = data)
V(graph)$color <- ifelse(data$d == relations_1$from[1], "red", "orange")
plot(graph, layout=layout.circle, edge.arrow.size = 0.2)
- But would there have been a way to "automatically" connect all the points in the graph directly using the "relations" data frame without manually creating a new data frame "relations_1"? Could a single line (or a few lines) of code have been added that would have automatically taken the "relations" data frame and connected everything together?
Thank you!
CodePudding user response:
You could just update relations using complete, and than filter out the rows where from is equal to to, which gives arrows from a node to itself.
relations <- relations %>%
complete(from, to) %>%
dplyr::filter(from != to)
CodePudding user response:
An solution using igraph::make_full_graph:
relations %>%
with(union(from,to)) %>%
{
G <- make_full_graph(length(.), directed = TRUE)
V(G)$name <- .
G
}