I am working with the R programming language.
Suppose I have the following function:
library(GA)
Rastrigin <- function(x1, x2)
{
20 x1^2 x2^2 - 10*(cos(2*pi*x1) cos(2*pi*x2))
}
As an example, suppose I try to optimize this function using some optimization algorithm and request for a very large number of iterations:
GA <- ga(type = "real-valued",
fitness = function(x) -Rastrigin(x[1], x[2]),
lower = c(-5.12, -5.12), upper = c(5.12, 5.12),
popSize = 50, maxiter = 10000, run = 10000)
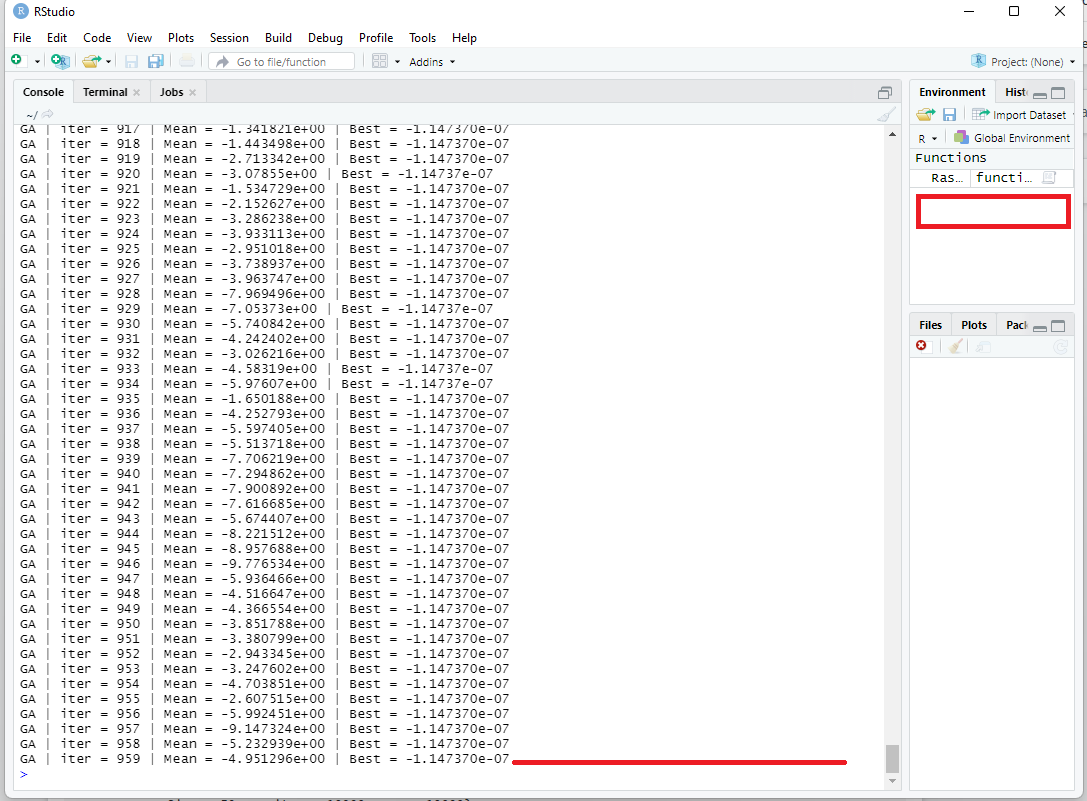
After running this code, I quickly clicked the "Red Stop Sign Button" to abort this code:
As seen here:
I requested the optimization algorithm to run for 10000 iterations, but I manually aborted the code after 959 iterations.
As a result, the "GA object" (i.e. the R object that stores the progress of the optimization) was never created.
GA
Error: object 'GA' not found
My Question: The computer has still performed a significant amount of work - ideally, I would like to see how far the computer reached with the optimization and inspect the most current solution obtained before the code was aborted. But since the "GA object" was never created, I can not extrapolate what the most current solution was from the log - I can only see the "mean" and "best" value of the function evaluated at the most current solution (and the not the current solution itself).
For example, suppose I let the optimization run to completion - I can then access the entire results of the optimization:
GA_complete <- ga(type = "real-valued",
fitness = function(x) -Rastrigin(x[1], x[2]),
lower = c(-5.12, -5.12), upper = c(5.12, 5.12),
popSize = 50, maxiter = 10, run = 10)
head(GA_complete@population)
[,1] [,2]
[1,] -0.04222106 -0.04219359
[2,] 0.33339796 -0.03329398
[3,] 0.59785640 -0.31436130
[4,] -0.07623396 0.15844942
[5,] 0.96052009 -0.04702174
[6,] 0.02373485 -0.03466375
head(GA_complete@fitness)
[1] -0.7027423 -15.3337890 -32.5594724 -5.7160470 -1.6641805 -0.3490039
Is it possible to somehow "access" the most current solution when the code is aborted? Can we somehow access the log and view the intermediate results/progress of aborted code?
Thanks!
CodePudding user response:
The ability to save work in the middle of large-scale processing relies wholly on the function itself, which means that if you want to have an interruptible call and preserve the interim results, then you need to rewrite the function itself so that it has this, will catch errors gracefully, and return data. One thing that would advise this is https://community.rstudio.com/t/how-to-catch-the-keyboard-interruption-in-r/7336, where one can use tryCatch with the interrupt= handler, as in:
tryCatch(for (i in 1:1e6) mean(rcauchy(1:1e6)), interrupt = function(e) "foo")
I think the conventional use of tryCatch is to catch error= and possibly warning=, but the use of interrupt= is not as often seen in the wild. (I've not looked for it ... but I've never had a function react gracefully to me interrupting it. I do know that some purrr:: functions use it.)
If you're not willing to invest the time to rewrite ga's internals in order to handle this (perhaps not a trivial undertaking), I suggest you handle it externally. Instead of running it 10000 times, perhaps you only run it 100 or 10 or so, and externally repeat it to fill to the desired volume.
Here's a helper function that might work for you. (I've not tested it with ga, but I imagine it can still be adapted to work with that.)
interruptible <- function(expr, n = 1) {
stopifnot(n > 0)
n <- as.integer(n)
expr <- substitute(expr)
out <- list()
interrupted <- structure(list(), class = "interrupted")
i <- 0L
while (i < n) {
this <- tryCatch(eval(expr), interrupt = function(e) interrupted)
if (inherits(this, "interrupted")) break
out <- c(out, list(this))
i <- i 1L
}
if (i < n) warning(sprintf("interrupted, %i/%i completed", i, n), call. = FALSE)
out
}
And a simple test-run:
out <- interruptible({Sys.sleep(2); message(Sys.time()); runif(1);}, 3)
# 2022-04-02 13:55:46
# 2022-04-02 13:55:48
# 2022-04-02 13:55:50
out
# [[1]]
# [1] 0.2934904
# [[2]]
# [1] 0.7578942
# [[3]]
# [1] 0.8208736
Let's interrupt that mid-run:
out <- interruptible({Sys.sleep(2); message(Sys.time()); runif(1);}, 3)
# 2022-04-02 13:55:55
# 2022-04-02 13:55:57
C-c C-c # <--- I use emacs/ess, this is me interrupting it
# Warning: interrupted, 1/3 completed
out
# [[1]]
# [1] 0.7229554
# [[2]]
# [1] 0.4721477
It is up to the caller to determine how to combine the results. In this case, it might simply be to concatenate the results (unlist or do.call(c, out)), but if a frame or matrix is returned, then it might be combined using do.call(rbind, out) or dplyr::bind_rows(out) or data.table::rbindlist(out).
I suspect (without testing) that you might be able to do something like:
GA_complete <- interruptible(
ga(type = "real-valued",
fitness = function(x) -Rastrigin(x[1], x[2]),
lower = c(-5.12, -5.12), upper = c(5.12, 5.12),
popSize = 50, maxiter = 10, run = 10),
n = 1000)
and then find a way to combine each of the results; since the return value is not a simple frame, it might be a little more elbow-grease to combine each of the @-slots, or perhaps you should not try to combine all of them, just the specific results you need.
N.B.: I recognize that running one genetic algorithm for 10 generations and repeating 1000 times is not the same as one single 10000-generation run. Unfortunately, in order to get that effect while being able to interrupt it with a saved-state, you will need to alter ga itself, and that I'm not willing to just "attempt" for an SO question. It is further complicated by the fact that ga can run in parallel.