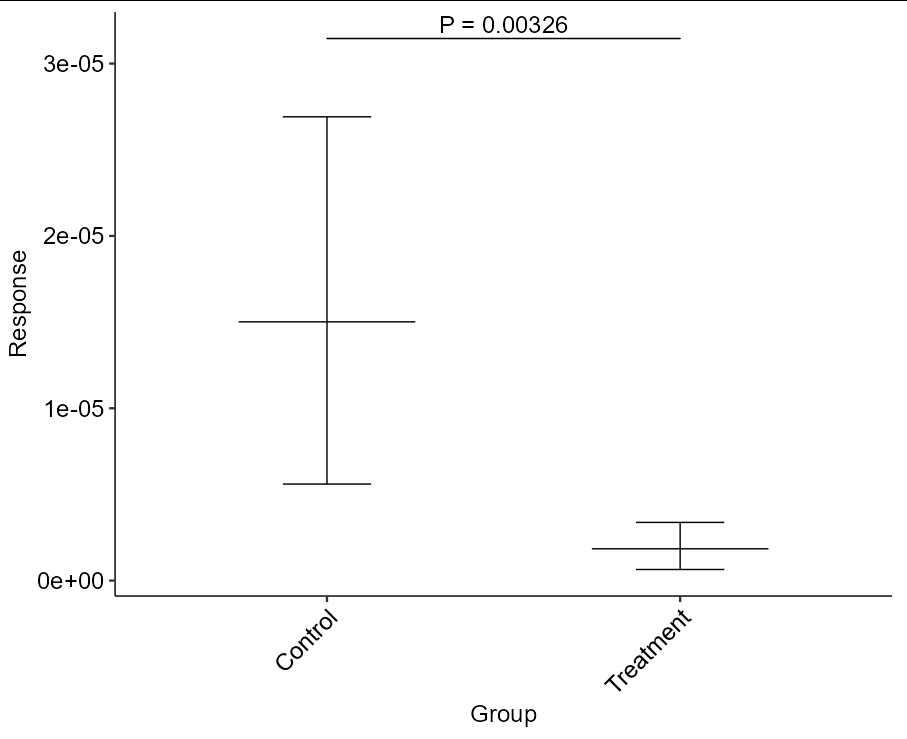
After performing an rstatix::wilcox_test() on my data I automatically added the y.position value for plotting the p value using rstatix::add_xy_position(). I then plotted the data and added the p value using ggpubr::stat_pvalue_manual(). The problem is that the y.position value is defined incorrectly as zero, causing the plotted p value and bracket geom to appear at the bottom of the y-axis at zero when it should appear near the top of the y-axis. The documentation for rstatix indicates that the y.position value, as calculated by the max default setting, should be have been equal to max(data$Response) = 0.00008556344, not zero. Am I using these functions incorrectly? If not, how can I get the p value and bracket geom to appear at the y-axis data value corresponding to the maximum?
library(tidyverse)
library(rstatix)
library(ggpubr)
# minimal data example
data <- tibble::tribble(
~Group, ~Response,
"Treatment", 3.210486e-06,
"Control", 4.006825e-06,
"Treatment", 4.350836e-06,
"Control", 4.216934e-06,
"Treatment", 4.415194e-06,
"Control", 1.0260606e-05,
"Treatment", 1.111064e-06,
"Control", 1.0779088e-05,
"Treatment", 3.57185e-07,
"Control", 1.139097e-06,
"Treatment", 0,
"Control", 2.31074e-07,
"Treatment", 5.78956e-07,
"Control", 4.371157e-06,
"Treatment", 6.5825e-08,
"Control", 9.587202e-06,
"Treatment", 2.65383e-07,
"Control", 3.57337e-06,
"Treatment", 7.14146e-07,
"Control", 3.868605e-06,
"Treatment", 1.2213951e-05,
"Control", 6.936899e-06,
"Treatment", 4.71707e-07,
"Control", 5.5957173e-05,
"Treatment", 1.265942e-06,
"Control", 8.5563441e-05,
"Treatment", 0,
"Control", 0,
"Treatment", 0,
"Control", 2.1306289e-05,
"Treatment", 5.2055e-07,
"Control", 1.8420094e-05
)
# performing Wilcoxon test and adding 'y.position' for the p value
wilcoxon_result <- data %>%
wilcox_test(Response ~ Group) %>%
add_xy_position("Group")
# the 'y.position' is incorrectly defined as zero.
wilcoxon_result
#> # A tibble: 1 × 11
#> .y. group1 group2 n1 n2 statistic p y.position groups xmin
#> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <name> <dbl>
#> 1 Response Control Treatm… 16 16 206. 0.00326 0 <chr> 1
#> # … with 1 more variable: xmax <dbl>
# the p value and bracket geom are plotted at zero.
ggplot(data, aes(Group, Response))
stat_summary(geom = "errorbar", fun.data = mean_cl_boot, width = 0.25)
stat_summary(geom = "crossbar", fun = mean, fun.min = mean, fun.max = mean, fatten = FALSE, width = 0.5)
stat_pvalue_manual(wilcoxon_result, "P = {p}", size = 18 / ggplot2::.pt, tip.length = 0, bracket.size = 2 / ggplot2::.stroke)
theme_pubr(base_size = 18, x.text.angle = 45)
CodePudding user response:
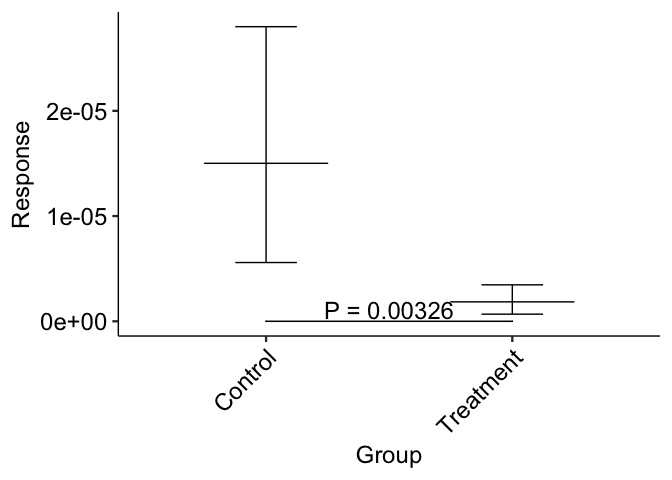
It seems your y position is being rounded to zero because you are dealing with very small numbers. If you multiply the Response variable by 1 million you get the same p-value, and you can then divide the y position to get it to sit wherever you want :
# performing Wilcoxon test and adding 'y.position' for the p value
wilcoxon_result <- data %>%
mutate(Response = Response * 1e6) %>%
wilcox_test(Response ~ Group) %>%
add_xy_position("Group") %>%
mutate(y.position = y.position /3e6)
This gives:
wilcoxon_result
#> # A tibble: 1 x 11
#> .y. group1 group2 n1 n2 statistic p y.position groups xmin xmax
#> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl> <named list> <dbl> <dbl>
#> Response Control Treatment 16 16 206. 0.00326 0.0000315 <chr [2]> 1 2
And the plot looks like this:
ggplot(data, aes(Group, Response))
stat_summary(geom = "errorbar", fun.data = mean_cl_boot, width = 0.25)
stat_summary(geom = "crossbar", fun = mean, fun.min = mean,
fun.max = mean, fatten = FALSE, width = 0.5)
stat_pvalue_manual(wilcoxon_result, "P = {p}",
size = 18 / ggplot2::.pt, tip.length = 0,
bracket.size = 2 / ggplot2::.stroke)
theme_pubr(base_size = 18, x.text.angle = 45)