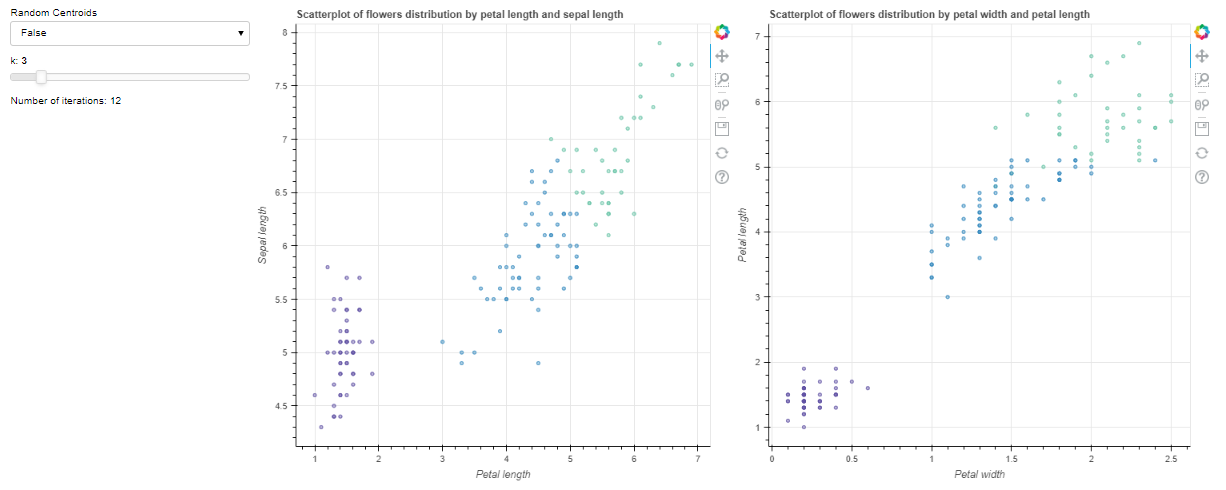
My intial plot works great and shows the colors of the intial 3 clusters. However, when I select a new k value with the silder widget, the new clusters are all colored grey.
import numpy as np
import pandas as pd
import random
from typing import List, Tuple
from bokeh.models import ColumnDataSource, Slider, Div, Select
from bokeh.sampledata.iris import flowers
from bokeh.plotting import figure, curdoc
from bokeh.layouts import column, row
from bokeh.palettes import Spectral10
from bokeh.transform import factor_cmap
# Use these centroids in the first iteration of you algorithm if "Random Centroids" is set to False in the Dashboard
DEFAULT_CENTROIDS = np.array([[5.664705882352942, 3.0352941176470587, 3.3352941176470585, 1.0176470588235293],
[5.446153846153847, 3.2538461538461543, 2.9538461538461536, 0.8846153846153846],
[5.906666666666667, 2.933333333333333, 4.1000000000000005, 1.3866666666666667],
[5.992307692307692, 3.0230769230769234, 4.076923076923077, 1.3461538461538463],
[5.747619047619048, 3.0714285714285716, 3.6238095238095243, 1.1380952380952383],
[6.161538461538462, 3.030769230769231, 4.484615384615385, 1.5307692307692309],
[6.294117647058823, 2.9764705882352938, 4.494117647058823, 1.4],
[5.853846153846154, 3.215384615384615, 3.730769230769231, 1.2076923076923078],
[5.52857142857143, 3.142857142857143, 3.107142857142857, 1.007142857142857],
[5.828571428571429, 2.9357142857142855, 3.664285714285714, 1.1]])
def get_closest(data_point: np.ndarray, centroids: np.ndarray):
"""
Takes a data_point and a nd.array of multiple centroids and returns the index of the centroid closest to data_point
by computing the euclidean distance for each centroid and picking the closest.
"""
N = centroids.shape[0]
dist = np.empty(N)
for i, c in enumerate(centroids):
dist[i] = np.linalg.norm(c - data_point)
index_min = np.argmin(dist)
return index_min
def to_classes(clustering):
# Get number of samples (you can pass it directly to the function)
num_samples = sum(x.shape[0] for x in clustering)
indices = np.empty((num_samples,)) # An empty array with correct size
for ith, cluster in enumerate(clustering):
# use cluster indices to assign to correct the cluster index
indices[cluster] = ith
return indices.astype(int)
def k_means(data_np: np.ndarray, k:int=3, n_iter:int=500, random_initialization=False) -> Tuple[np.ndarray, int]:
"""
:param data: your data, a numpy array with shape (n_entries, n_features)
:param k: The number of clusters to compute
:param n_iter: The maximal numnber of iterations
:param random_initialization: If False, DEFAULT_CENTROIDS are used as the centroids of the first iteration.
:return: A tuple (cluster_indices: A numpy array of cluster_indices,
n_iterations: the number of iterations it took until the algorithm terminated)
"""
# Initialize the algorithm by assigning random cluster labels to each entry in your dataset
k=k 1
centroids = data_np[random.sample(range(len(data_np)), k)]
labels = np.array([np.argmin([(el - c) ** 2 for c in centroids]) for el in data_np])
clustering = []
for k in range(k):
clustering.append(data_np[labels == k])
# Implement K-Means with a while loop, which terminates either if the centroids don't move anymore, or
# if the number of iterations exceeds n_iter
counter = 0
while counter < n_iter:
# Compute the new centroids, if random_initialization is false use DEFAULT_CENTROIDS in the first iteration
# if you use DEFAULT_CENTROIDS, make sure to only pick the k first entries from them.
if random_initialization is False and counter == 0:
centroids = DEFAULT_CENTROIDS[random.sample(range(len(DEFAULT_CENTROIDS)), k)]
# Update the cluster labels using get_closest
labels = np.array([get_closest(el, centroids) for el in data_np])
clustering = []
for i in range(k):
clustering.append(np.where(labels == i)[0])
counter = 1
new_centroids = np.zeros_like(centroids)
for i in range(k):
if len(clustering[i]) > 0:
new_centroids[i] = data_np[clustering[i]].mean(axis=0)
else:
new_centroids[i] = centroids[i]
# if the centroids didn't move, exit the while loop
if clustering is not None and (centroids != new_centroids).sum() == 0:
break
else:
centroids = new_centroids
pass
# return the final cluster labels and the number of iterations it took
clustering = to_classes(clustering)
return clustering, counter
def callback(attr, old, new):
# recompute the clustering and update the colors of the data points based on the result
k = slider_k.value_throttled
init = select_init.value
clustering_new, counter_new = k_means(data_np,k,500,init)
source.data.update((ColumnDataSource(dict(petal_length=data['petal_length'],
sepal_length=data['sepal_length'],
petal_width=data['petal_width'],
clustering=clustering_new.astype(str)))).data)
div.text = 'Number of iterations: %d'%(counter_new)
pass
# read and store the dataset
data: pd.DataFrame = flowers.copy(deep=True)
data = data.drop(['species'], axis=1)
# Create a copy of the data as numpy array, which you can use for computing the clustering
data_np = np.asarray(data)
# Create the dashboard
# 1. A Select widget to choose between random initialization or using the DEFAULT_CENTROIDS on top
select_init = Select(title='Random Centroids',value='False',options=['True','False'])
# 2. A Slider to choose a k between 2 and 10 (k being the number of clusters)
slider_k = Slider(start=2,end=10,value=3,step=1,title='k')
# 4. Connect both widgets to the callback
select_init.on_change('value',callback)
slider_k.on_change('value_throttled',callback)
# 3. A ColumnDataSource to hold the data and the color of each point you need
clustering, counter = k_means(data_np,3,500,False)
source = ColumnDataSource(dict(petal_length=data['petal_length'],
sepal_length=data['sepal_length'],
petal_width=data['petal_width'],
clustering=clustering.astype(str)))
# 4. Two plots displaying the dataset based on the following table, have a look at the images
# in the handout if this confuses you.
#
# Axis/Plot Plot1 Plot2
# X Petal length Petal width
# Y Sepal length Petal length
#
# Use a categorical color mapping, such as Spectral10, have a look at this section of the bokeh docs:
# https://docs.bokeh.org/en/latest/docs/user_guide/categorical.html#filling
plot1 = figure(title='Scatterplot of flowers distribution by petal length and sepal length')
plot1.yaxis.axis_label = 'Sepal length'
plot1.xaxis.axis_label = 'Petal length'
plot1.scatter(x='petal_length',
y='sepal_length',
fill_alpha=0.4,
source=source,
color=factor_cmap('clustering',
palette=Spectral10,
factors=np.unique(clustering).astype(str)))
plot2 = figure(title='Scatterplot of flowers distribution by petal width and petal length')
plot2.yaxis.axis_label = 'Petal length'
plot2.xaxis.axis_label = 'Petal width'
plot2.scatter(x='petal_width',
y='petal_length',
fill_alpha=0.4,
source=source,
color=factor_cmap('clustering',
palette=Spectral10,
factors=np.unique(clustering).astype(str)))
# 5. A Div displaying the currently number of iterations it took the algorithm to update the plot.
div = Div(text='Number of iterations: %d'%(counter))
div.on_change('text',callback)
lt = row(column(select_init,slider_k,div),plot1,plot2)
curdoc().add_root(lt)
bokeh serve --show file_name.py
Then when I select a new k value with the slider, the graph looks like so.
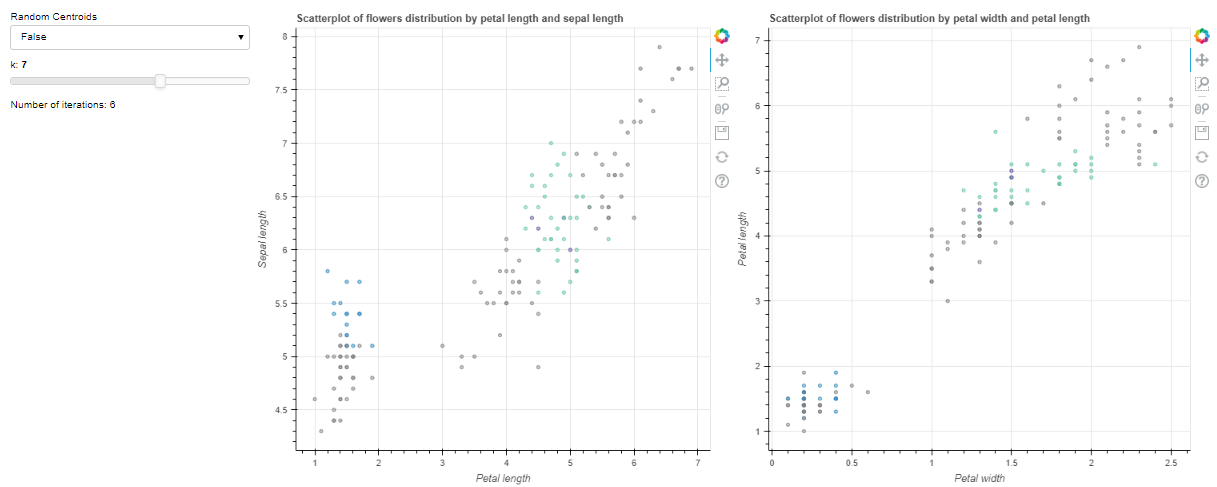
How can I update the colors of each cluster in the callback function?
This is the expected output after selecting k=7
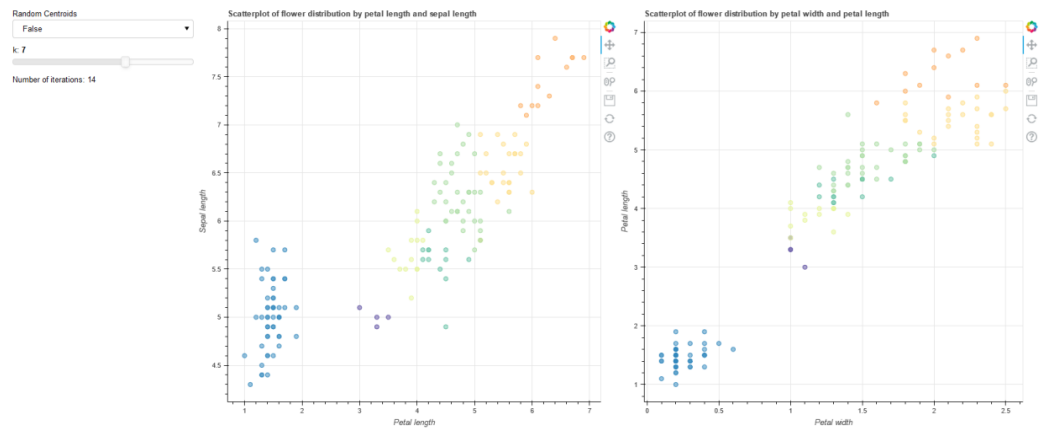
CodePudding user response:
This seemed to work in my callback function
def callback(attr, old, new):
# recompute the clustering and update the colors of the data points based on the result
k = slider_k.value_throttled
init = select_init.value
clustering_new, counter_new = k_means(data_np,k,500,init)
source.data['clustering'] = clustering_new.astype(str)
mapper = factor_cmap('clustering',palette=Spectral10,factors=np.unique(clustering_new).astype(str))
scatter1.glyph.fill_color = mapper
scatter2.glyph.fill_color = mapper
scatter1.glyph.line_color = mapper
scatter2.glyph.line_color = mapper
div.text = 'Number of iterations: %d'%(counter_new)
pass
And changed the arguments in my plots
mapper = factor_cmap('clustering',palette=Spectral10,factors=np.unique(clustering).astype(str))
plot1 = figure(title='Scatterplot of flowers distribution by petal length and sepal length')
plot1.yaxis.axis_label = 'Sepal length'
plot1.xaxis.axis_label = 'Petal length'
scatter1 = plot1.scatter(x='petal_length',
y='sepal_length',
fill_alpha=0.4,
source=source,
fill_color=mapper,
line_color=mapper)
plot2 = figure(title='Scatterplot of flowers distribution by petal width and petal length')
plot2.yaxis.axis_label = 'Petal length'
plot2.xaxis.axis_label = 'Petal width'
scatter2 = plot2.scatter(x='petal_width',
y='petal_length',
fill_alpha=0.4,
source=source,
fill_color=mapper,
line_color=mapper)