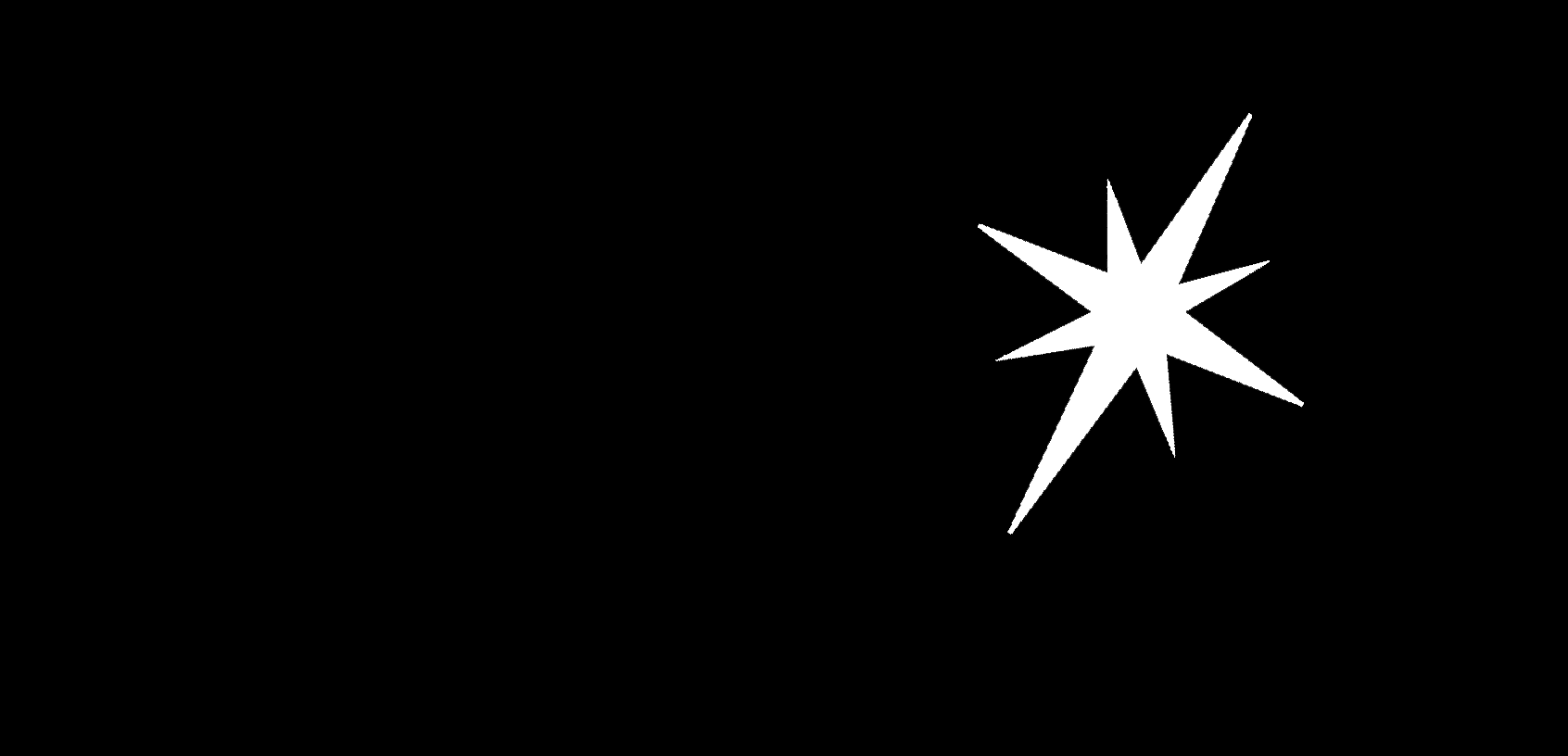
Need to run though an image in python and essentially calculate the average location of all acceptable pixels within a certain boundary. The image is black and white. The acceptable pixels have a value of 255 and the unacceptable pixels have a value of zero. The image is something like 2592x1944 and takes maybe 15 seconds to run. This will need to be looped several times. Is there a faster way to do this?
goodcount = 0
sumx=0
sumy=0
xindex=0
yindex=0
for row in mask:
yindex =1
xindex=0
for n in row:
xindex =1
if n == 255:
goodcount = 1
sumx = xindex
sumy = yindex
if goodcount != 0:
y = int(sumy / goodcount)
x = int(sumx / goodcount)
CodePudding user response:
np.where() will return arrays of the indices where a condition is true, which we can average (adding 1 to match your indexing) and cast to an integer:
if np.any(mask == 255):
y, x = [int(np.mean(indices 1)) for indices in np.where(mask == 255)]
CodePudding user response:
I think you are looking for the centroid of the white pixels, which OpenCV will find for you very fast:
#!/usr/bin/env python3
import cv2
import numpy as np
# Load image as greyscale
im = cv2.imread('star.png')
# Make greyscale version
grey = cv2.cvtColor(im, cv2.COLOR_BGR2GRAY)
# Ensure binary
_, grey = cv2.threshold(grey,127,255,0)
# Calculate moments
M = cv2.moments(grey)
# Calculate x,y coordinates of centre
cX = int(M["m10"] / M["m00"])
cY = int(M["m01"] / M["m00"])
# Mark centroid with circle, and tell user
cv2.circle(im, (cX, cY), 10, (0, 0, 255), -1)
print(f'Centroid at location: {cX},{cY}')
# Save output file
cv2.imwrite('result.png', im)
Sample Output
Centroid at location: 1224,344
That takes 381 microseconds on the 1692x816 image above, and rises to 1.38ms if I resize the image to the dimensions of your image... I'm calling that a 10,800x speedup