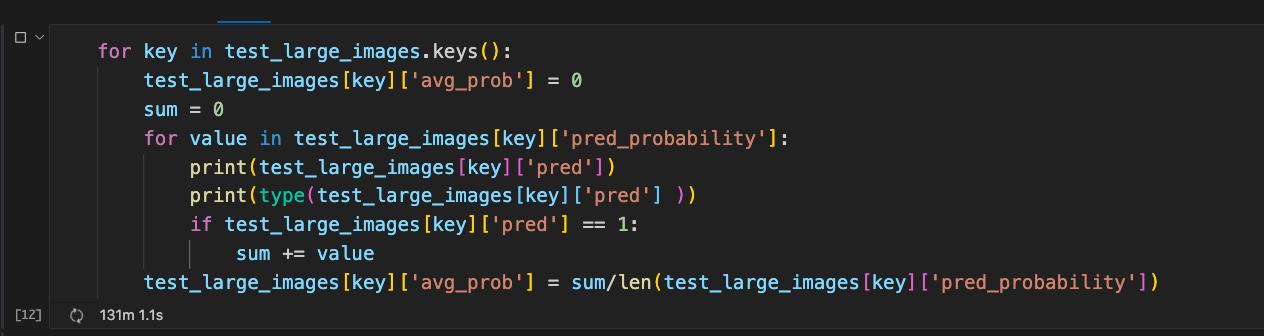
I have the following code:
for key in test_large_images.keys():
test_large_images[key]['avg_prob'] = 0
sum = 0
for value in test_large_images[key]['pred_probability']:
print(test_large_images[key]['pred'])
print(type(test_large_images[key]['pred'] ))
if test_large_images[key]['pred'] == 1:
sum = value
test_large_images[key]['avg_prob'] = sum/len(test_large_images[key]['pred_probability'])
and it is a dictionary of 359 large images that each could have between 200 to 8000 smaller images which I call patches. The test_large_images is a dictionary of inferences on the smaller images which also have prediction probability, large image name, patch name, etc for each patch. My goal is to find the average probability of the larger image based on the prediction probability of smaller patches prediction probabilities inside that image. When I run this loop on a smaller dataset (45K patches) for which I have saved its inferences in a pkl file, it runs very fast. But, it's been more than 130 minutes that this script has been running as you can see in Jupyter Notebook remotely on VSCode Remote (with a local client on Mac).
Is there a way I can take advantage of the 24 CPU cores I have to accelerate this nested dictionary computation?
CodePudding user response:
- Don't use
sumas a variable name, since it's a built in function. - The line
test_large_images[key]['avg_prob'] = 0is not needed. - PeterK is correct that your condition doesn't need to be calculated every time in the inner for loop.
- Why are we printing these repeatedly, or was that just for testing?
for key in test_large_images.keys():
add = 0
condition = test_large_images[key]['pred'] == 1 # This is what PeterK means by take it out (of the loop).
for value in test_large_images[key]['pred_probability']:
# print(test_large_images[key]['pred'])
# print(type(test_large_images[key]['pred']))
if condition:
add = value
test_large_images[key]['avg_prob'] = add/len(test_large_images[key]['pred_probability'])
Your code could be simplified to:
for key in test_large_images.keys():
condition = test_large_images[key]['pred'] == 1
num = sum(x for x in test_large_images[key]['pred_probability'] if condition)
denom = len(test_large_images[key]['pred_probability'])
test_large_images[key]['avg_prob'] = num/denom
Based off of feedback and some additional optimization:
for key in test_large_images.keys():
if test_large_images[key]['pred'] != 1:
test_large_images[key]['avg_prob'] = 0
continue
values = test_large_images[key]['pred_probability']
test_large_images[key]['avg_prob'] = sum(values)/len(values)
Here's two different types of averaging (and I am most interested in taking the average of probabilities only over the number of entries for which the prediction was 1). I call that avg_prob_pos
for key in progress_bar(test_large_images.keys()):
condition = test_large_images[key]['pred'] == 1
num = sum(x for x in test_large_images[key]['pred_probability'] if condition)
denom = len(test_large_images[key]['pred_probability'])
count = sum(x for x in test_large_images[key]['pred'] if condition)
if count != 0:
test_large_images[key]['avg_prob_pos'] = num/count
test_large_images[key]['avg_prob'] = num/denom
percentage = test_large_images[key]['pred'].count(1)/len(test_large_images[key]['pred'])
test_large_images[key]['percentage'] = percentage