A data file is imported to a SQL Server table. One of the columns in data file is of text data type with values in this column to be integers only. The corresponding column in destination table in SQL Server db is of type varchar(100). But after data import, SQL Server is storing the values such as 0474525431 as 4.74525431E8 that is a Scientific Notations.
Question: In the above situation how can we prevent SQL server from storing the values into Scientific Notations. For example when 0474525431 is inserted into a VARCHAR(100) column it should be stored as is and not as 4.74525431E8
UPDATE:
The code to import data:
from pyspark.sql.functions import *
df = spark.read.csv(".../Test/MyFile.csv", header="true", inferSchema="true")
server_name = "jdbc:sqlserver://{SERVER_ADDR}"
database_name = "database_name"
url = server_name ";" "databaseName=" database_name ";"
table_name = "table_name"
username = "username"
password = "myPassword"
try:
df.write \
.format("com.microsoft.sqlserver.jdbc.spark") \
.mode("overwrite") \
.option("url", url) \
.option("dbtable", table_name) \
.option("user", username) \
.option("password", password) \
.save()
except ValueError as error :
print("Connector write failed", error)
UPDATE2:
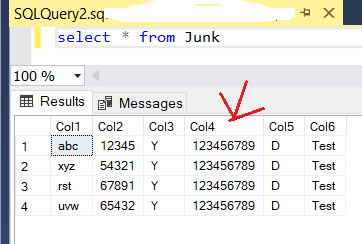
The issue seems to be related to the leading zeros, as well. I created a sample file (shown below) and imported its data to corresponding SQL table (also shown below) and noticed that the leading zeros were removed. This is happening despite the fact that all 6 columns in data file and the table are text (varchar):
Data file:
Col1|Col2|Col3|Col4|Col5|Col6
abc|12345|Y|0123456789|D|Test
xyz|54321|Y|0123456789|D|Test
rst|67891|Y|0123456789|D|Test
uvw|65432|Y|0123456789|D|Test
CodePudding user response:
Spark is inferring the schema, and it's picking integer for the column containing the value '0474525431'. So as the DataFrame is read the value is convered to an integer and the leading zero is discarded.
So you need to make sure the DataFrame has the correct types. You can either specify the schema explicitly when you create the DataFrame, or turn off inferSchema and then convert selected columns to different types before loading to SQL Server. Examples of both are in this answer.
Here's a sample showing that the leading '0' is preserved if the DataFrame uses a string column rather than an integer column.
from pyspark.sql.functions import *
jdbcUrl = "jdbc:sqlserver://{0}:{1};database={2}".format(jdbcHostname, jdbcPort, jdbcDatabase)
connectionProperties = {
"user" : jdbcUsername,
"password" : jdbcPassword,
"driver" : "com.microsoft.sqlserver.jdbc.SQLServerDriver"
}
pushdown_query = "(select '0474525431' f, * from sys.objects) emp"
df = spark.read.jdbc(url=jdbcUrl, table=pushdown_query, properties=connectionProperties)
df.write.mode("overwrite").option("header",True).csv("datalake/temp.csv")
df = spark.read.csv("datalake/temp.csv", header="true", inferSchema="false")
df.printSchema()
table_name = "table_name"
try:
df.write \
.format("com.microsoft.sqlserver.jdbc.spark") \
.mode("overwrite") \
.option("url", jdbcUrl) \
.option("dbtable", table_name) \
.option("user", jdbcUsername) \
.option("password", jdbcPassword) \
.save()
except ValueError as error :
print("Connector write failed", error)
df = spark.read.jdbc(url=jdbcUrl, table=table_name, properties=connectionProperties)
display(df)