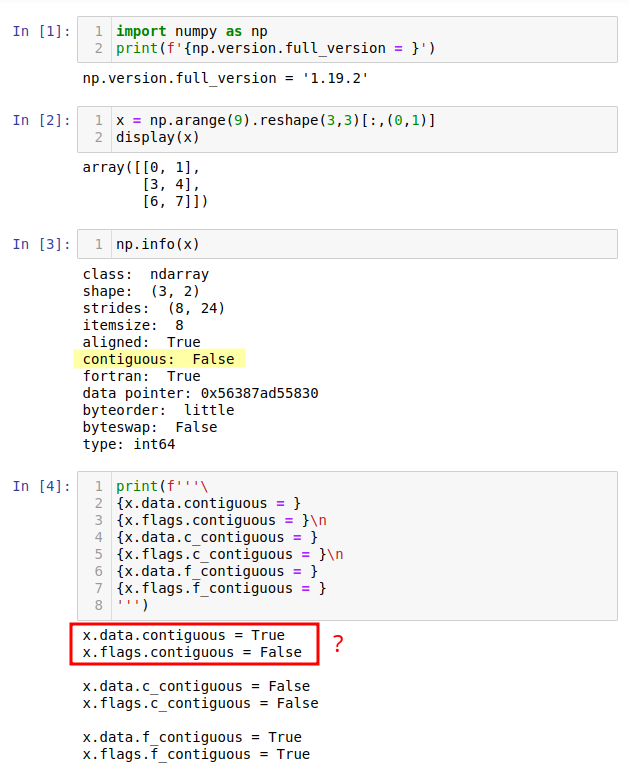
While experimenting with Numpy, I found that the contiguous value provided by numpy.info may differ from numpy.ndarray.data.contiguous (see the code and screenshot below).
import numpy as np
x = np.arange(9).reshape(3,3)[:,(0,1)]
np.info(x)
print(f'''
{x.data.contiguous = }
{x.flags.contiguous = }
{x.data.c_contiguous = }
{x.flags.c_contiguous = }
{x.data.f_contiguous = }
{x.flags.f_contiguous = }
''')
CodePudding user response:
x = np.arange(9).reshape(3,3)[:,(0,1)]
np.arange(9) produces a 1d array; reshape(3,3) reshapes it to 2d. It's a view of the original arange. Without an order parameter, reshape sticks with the default c-order.
The [0,[0,1]] is advanced indexing, making a copy. Indexing with [0,:2] would select the same values, but make a view.
The info strides is (8,24). The strides for x after the reshape should be (24,8), stepping by 8 bytes for the last dimension, 3*8 for first. But the advanced indexing flips things around - that's a detail of indexing that we usually ignore (or are unaware of).
2d array with smaller first strides is F-order.
I won't try to decipher all the data/flats contiguous prints, but the basic layout is obvious to me from the shape and strides. I think the strides has priority, and all the 'contiguous' displays are derivative, interpretations, so to speak, of the strides.
With 3d (or higher) arrays, the contiguous alternatives can break down. It would be possible to make an array with strides like (48,8,24), where the middle dimension steps the fastest. That's neither c or f congtiguous.
I might add that unless you are doing something like
np.arange(9).reshape(3,3, order='F')
the type of contiguity is usually not something we worry about. Some functions (esp. compiled ones) require a certain contiguity. And some operations are faster (or slower) depending on which dimension is 'inner-most'. But for ordinary numpy use I don't pay much attention to the flags. I used numpy for years before realizing that your example indexing flipped the order.
You could have just displayed x.flags. I'm not sure what displaying x.data does for you.
CodePudding user response:
Let's see how numpy.info works. From the source code we can see the subroutine for processing ndarray:
def info(object=None, maxwidth=76, output=None, toplevel='numpy'):
...
elif isinstance(object, ndarray):
_info(object, output=output)
...
def _info(obj, output=None):
"""Provide information about ndarray obj"""
bp = lambda x: x
...
print("contiguous: ", bp(obj.flags.contiguous), file=output)
print("fortran: ", obj.flags.fortran, file=output)
...
It returns flags.contiguous as the array's continuity parameter. This one isn't specified in flags description. But we can find it in flagsobject.c:
// ...
static PyGetSetDef arrayflags_getsets[] = {
{"contiguous",
(getter)arrayflags_contiguous_get,
NULL,
NULL, NULL},
{"c_contiguous",
(getter)arrayflags_contiguous_get,
NULL,
NULL, NULL},
// ...
We can see here that a contiguous parameter from numpy.info is actually flags.c_contiguous and has nothing in common with ndarray.data.contiguous. I guess when programming in C it was natural to say just contiguous instead of c_contiguous, and this has led to a slight inconsistency in terminology.