I have a function that lists all the accessible environments in the current session.
env_list <- function(start_env) {
# convenience function
last <- function(s) { s[[length(s)]]}
res <- list(start_env)
while(!identical(emptyenv(), last(res))) {
res <- c(res, list(parent.env(last(res))))
}
res
}
env_list(globalenv())
The result i'm getting looks something like this:
[[1]]
<environment: R_GlobalEnv>
[[2]]
<environment: package:rlang>
attr(,"name")
[1] "package:rlang"
attr(,"path")
[1] "C:/Users/Documents/R/win-library/4.1/rlang"
[[3]]
<environment: package:igraph>
attr(,"name")
[1] "package:igraph"
attr(,"path")
[1] "C:/Users/Documents/R/win-library/4.1/igraph"
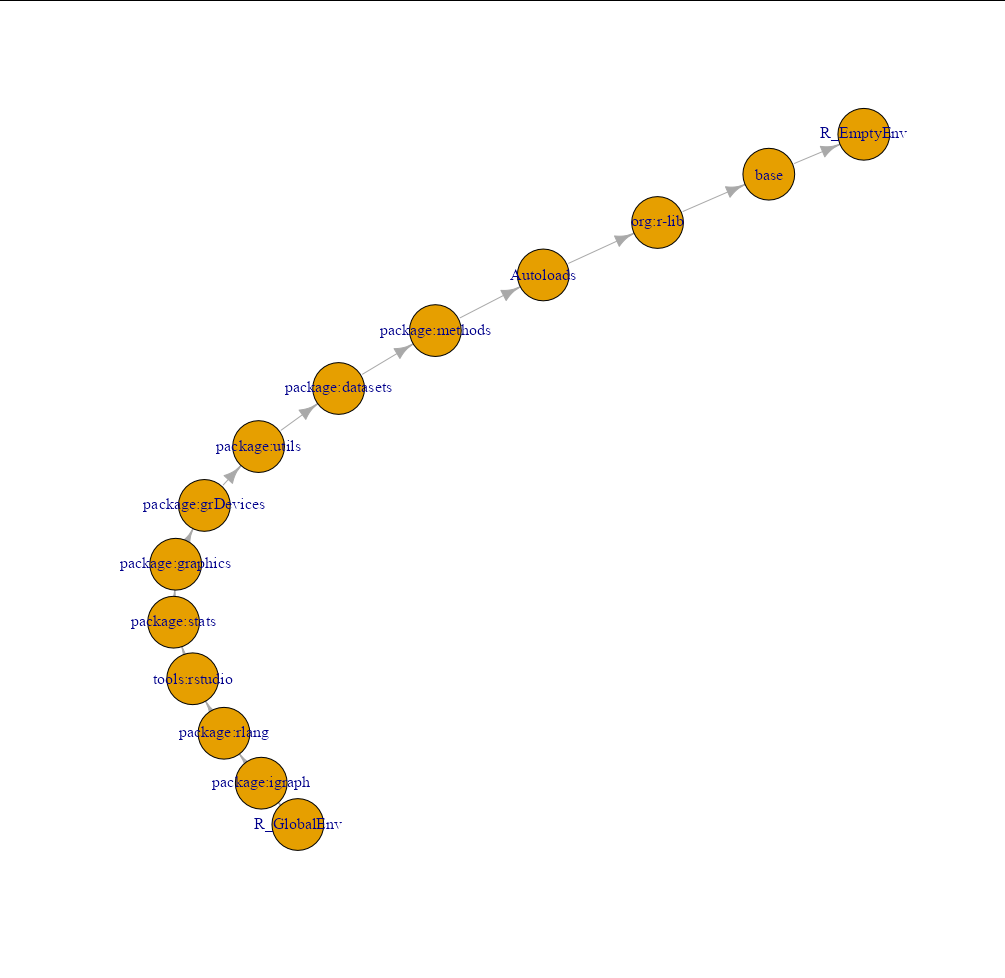
Could you advise approach to convert this list to a graph? I want to plot it with igraph later. Elements 1, 2, 3 probably should be nodes, but then what is the best way to denote edges, connecting parent and child environments?
CodePudding user response:
The obvious way to do this is to create an edgelist as a two-column character matrix with each environment's name in the left column, and its parent's name in the right column:
library(igraph)
my_envs <- env_list(globalenv())
edgelist <- t(sapply(my_envs[-length(my_envs)], function(x) {
c(environmentName(x), environmentName(parent.env(x)))
}))
g <- graph_from_edgelist(edgelist)
plot(g)