I am trying to use Numba for the first time in colab and I think I have successfully installed Numba since the @jit is not undefined now but I am getting errors in my code. The following is my attempt:
!apt-get install nvidia-cuda-toolkit
!pip3 install numba
import os
dev_lib_path = !find / -iname 'libdevice'
nvvm_lib_path = !find / -iname 'libnvvm.so'
assert len(dev_lib_path)>0, "Device Lib Missing"
assert len(nvvm_lib_path)>0, "NVVM Missing"
os.environ['NUMBAPRO_LIBDEVICE'] = dev_lib_path[0]
os.environ['NUMBAPRO_NVVM'] = nvvm_lib_path[0]
import numpy as np
import matplotlib.pyplot as plt
import random
from math import *
from random import *
from numba import jit
n=1000
mu=np.random.uniform(0,1,n)
r=[sqrt(-2*log(1-i)) for i in mu]
eta=np.random.uniform(0,1,n)
theta=2*pi*eta;
cuz=[cos(i) for i in theta]
suz=[sin(i) for i in theta]
Zinitial=[a*b for a,b in zip(r,cuz)];
Pinitial=[a*b for a,b in zip(r,suz)];
class Particle:
def __init__(self, pos, mom, spin):
self.pos = pos
self.mom = mom
self.spin = spin
SP = sorted(np.array([Particle(pos = i, mom = j, spin = choice([1, 0])) for i,j in zip(Zinitial,Pinitial)]),key=lambda x:x.pos)
Upi=[];
Downi=[];
count_plot=[];
for j in range(len(SP)):
if SP[j].spin == 1:
Upi.append(SP[j].pos)
else:
Downi.append(SP[j].pos)
Zavgi=sum(Zinitial)/len(Zinitial)
Zreli=sum(Upi)/len(Upi)-sum(Downi)/len(Downi)
"Observables"
"Time"
iter=10**(5);
dt=1/(2*n);
alf=sqrt(n);
"Dynamics"
@jit(nopython=True)
def f():
counter=0;
sum1,sum2=0,0;
Zavg=[Zavgi];
Zrelm=[Zreli];
T_plot=[0];
for i in range(1,iter 1):
t=i*dt;
T_plot.append(t)
Z=[];
Up=[];
Down=[];
c,s=cos(t),sin(t);
c1,s1=cos(t-dt),sin(t-dt);
for j in range(n-1):
collchk=((c*(SP[j].pos) s*(SP[j].mom))-(c*(SP[j 1].pos) s*(SP[j 1].mom)))*(c1*(SP[j].pos) s1*(SP[j].mom)-(c1*(SP[j 1].pos) s1*(SP[j 1].mom)));
prel=((c*(SP[j].mom)-s*(SP[j].pos))-(c*(SP[j 1].mom)-s*(SP[j 1].pos)))/2;
rcoeff=1/(1 (prel*alf)**2);
rand_value=random();
if collchk<0:
SP[j], SP[j 1]=SP[j 1],SP[j];
if rcoeff>rand_value:
counter=counter 1
SP[j].spin,SP[j 1].spin=SP[j 1].spin,SP[j].spin;
if SP[j].spin == 1:
Up.append(c*(SP[j].pos) s*(SP[j].mom))
else:
Down.append(c*(SP[j].pos) s*(SP[j].mom))
Z.append(c*(SP[j].pos) s*(SP[j].mom))
Zrel=sum(Up[0:])/len(Up) - sum(Down[0:])/len(Down);
Zrelm.append(Zrel)
Zm=sum(Z)/len(Z)
Zavg.append(Zm)
return [Zavg, Zrelm, counter]
Th error that I get:
TypingError: Failed in nopython mode pipeline (step: nopython frontend)
Untyped global name 'SP': Cannot type list element of <class '__main__.Particle'>
File "<ipython-input-12-45468d2d0903>", line 65:
def f(iter):
<source elided>
for j in range(n-1):
collchk=((c*(SP[j].pos) s*(SP[j].mom))-(c*(SP[j 1].pos) s*(SP[j 1].mom)))*(c1*(SP[j].pos) s1*(SP[j].mom)-(c1*(SP[j 1].pos) s1*(SP[j 1].mom)));
In the end I want to plot the lists that I have returned. Any help would be appreciated and if there is a way to use Numba even for the class definiton that would be great.
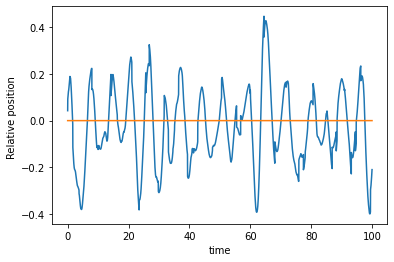
EDIT: After adding definition of sum in @Vandan code given below I get the following graph:
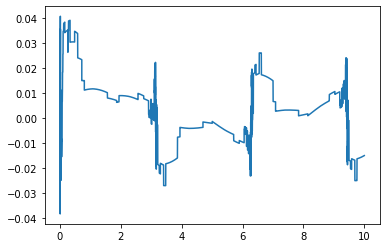
This should be a smoothly oscillating function like below:
I think the problem is because of the Numba which is added
import numpy as np
import matplotlib.pyplot as plt
import random
from math import *
from random import *
from numba import jit
@jit(nopython=True)
def sum(list2):
a=0;
for i in range(len(list2)):
a=a list2[i];
return a
"Dynamics"
@jit(nopython=True)
def f(SP, Zavgi, Zreli, alf, dt, n):
"Time"
counter = 0;
sum1, sum2 = 0, 0;
Zavg = np.array([Zavgi]);
Zrelm = np.array([Zreli]);
T_plot = [0];
for i in range(1, iter 1):
t = i * dt;
T_plot.append(t)
Z = [];
Up = [];
Down = [];
c, s = cos(t), sin(t);
c1, s1 = cos(t - dt), sin(t - dt);
for j in range(n - 1):
collchk = ((c * (SP[j][0]) s * (SP[j][1])) - (c * (SP[j 1][0]) s * (SP[j 1][1]))) * (
c1 * (SP[j][0]) s1 * (SP[j][1]) - (c1 * (SP[j 1][0]) s1 * (SP[j 1][1])));
prel = ((c * (SP[j][1]) - s * (SP[j][0])) - (c * (SP[j 1][1]) - s * (SP[j 1][0]))) / 2;
rcoeff = 1 / (1 (prel * alf) ** 2);
rand_value = random();
if collchk < 0:
SP[j], SP[j 1] = SP[j 1], SP[j];
if rcoeff > rand_value:
counter = counter 1
SP[j][2], SP[j 1][2] = SP[j 1][2], SP[j][2];
if SP[j][2] == 1:
Up.append(c * (SP[j][0]) s * (SP[j][1]))
else:
Down.append(c * (SP[j][0]) s * (SP[j][1]))
Z.append(c * (SP[j][0]) s * (SP[j][1]))
Zrel = sum(Up[0:]) / len(Up) - sum(Down[0:]) / len(Down);
Zrelm = np.append(Zrelm, Zrel)
Zm = sum(Z) / len(Z)
Zavg = np.append(Zavg, Zm)
return Zavg, Zrelm, counter,T_plot
if __name__ == '__main__':
n = 1000
mu = np.random.uniform(0, 1, n)
r = [sqrt(-2 * log(1 - i)) for i in mu]
eta = np.random.uniform(0, 1, n)
theta = 2 * pi * eta;
cuz = [cos(i) for i in theta]
suz = [sin(i) for i in theta]
Zinitial = [a * b for a, b in zip(r, cuz)];
Pinitial = [a * b for a, b in zip(r, suz)];
iter = 10 ** (6);
dt = 1 / (100 * n);
alf = sqrt(n);
SP = np.array(sorted(np.array([ np.array([i,j,choice([0,1])]) for i, j in zip(Zinitial, Pinitial)]),
key=lambda x: x[0]))
Upi = [];
Downi = [];
count_plot = [];
for j in range(len(SP)):
if SP[j][2] == 1:
Upi.append(SP[j][0])
else:
Downi.append(SP[j][0])
Zavgi = sum(Zinitial) / len(Zinitial)
Zreli = sum(Upi) / len(Upi) - sum(Downi) / len(Downi)
Zavg, Zrelm, counter,T_plot = f(SP, Zavgi, Zreli, alf, dt, n)
plt.plot(T_plot, Zrelm)
CodePudding user response:
I modified your code a bit and was able to get the function to run. I removed the Particle class and changed all the list instances to numpy arrays.
This is the output for the Zavg, Zrelm and counter:
Zavg: [0.07047501 0.06735052 0.06728123 ... 0.39516435 0.3947497 0.39433495]
Zrelm: [-0.04179043 -0.04461464 -0.0394889 ... -0.11080628 -0.11087257
-0.11093883]
Counter: 538
Here is the modified code:
import numpy as np
import matplotlib.pyplot as plt
import random
from math import *
from random import *
from numba import jit
"Dynamics"
@jit(nopython=True)
def f(SP, Zavgi, Zreli, alf, dt, n):
"Time"
counter = 0;
sum1, sum2 = 0, 0;
Zavg = np.array([Zavgi]);
Zrelm = np.array([Zreli]);
T_plot = [0];
for i in range(1, iter 1):
t = i * dt;
T_plot.append(t)
Z = [];
Up = [];
Down = [];
c, s = cos(t), sin(t);
c1, s1 = cos(t - dt), sin(t - dt);
for j in range(n - 1):
collchk = ((c * (SP[j][0]) s * (SP[j][1])) - (c * (SP[j 1][0]) s * (SP[j 1][1]))) * (
c1 * (SP[j][0]) s1 * (SP[j][1]) - (c1 * (SP[j 1][0]) s1 * (SP[j 1][1])));
prel = ((c * (SP[j][1]) - s * (SP[j][0])) - (c * (SP[j 1][1]) - s * (SP[j 1][0]))) / 2;
rcoeff = 1 / (1 (prel * alf) ** 2);
rand_value = random();
if collchk < 0:
SP[j], SP[j 1] = SP[j 1], SP[j];
if rcoeff > rand_value:
counter = counter 1
SP[j][2], SP[j 1][2] = SP[j 1][2], SP[j][2];
if SP[j][2] == 1:
Up.append(c * (SP[j][0]) s * (SP[j][1]))
else:
Down.append(c * (SP[j][0]) s * (SP[j][1]))
Z.append(c * (SP[j][0]) s * (SP[j][1]))
Zrel = sum(Up[0:]) / len(Up) - sum(Down[0:]) / len(Down);
Zrelm = np.append(Zrelm, Zrel)
Zm = sum(Z) / len(Z)
Zavg = np.append(Zavg, Zm)
return Zavg, Zrelm, counter
if __name__ == '__main__':
n = 1000
mu = np.random.uniform(0, 1, n)
r = [sqrt(-2 * log(1 - i)) for i in mu]
eta = np.random.uniform(0, 1, n)
theta = 2 * pi * eta;
cuz = [cos(i) for i in theta]
suz = [sin(i) for i in theta]
Zinitial = [a * b for a, b in zip(r, cuz)];
Pinitial = [a * b for a, b in zip(r, suz)];
iter = 10 ** (5);
dt = 1 / (2 * n);
alf = sqrt(n);
SP = np.array(sorted(np.array([ np.array([i,j,choice([0,1])]) for i, j in zip(Zinitial, Pinitial)]),
key=lambda x: x[0]))
Upi = [];
Downi = [];
count_plot = [];
for j in range(len(SP)):
if SP[j][2] == 1:
Upi.append(SP[j][0])
else:
Downi.append(SP[j][0])
Zavgi = sum(Zinitial) / len(Zinitial)
Zreli = sum(Upi) / len(Upi) - sum(Downi) / len(Downi)
Zavg, Zrelm, counter = f(SP, Zavgi, Zreli, alf, dt, n)
print(Zavg, Zrelm, counter)
CodePudding user response:
The error appears because Numba try to access to global variable which the type is not known at compile time. Indeed, SP is a pure-Python list called reflected list which can contain items of different types. Numba does not support such kind of list anymore. Instead, Numba supports typed lists which are compatible with reflected lists. This means you need to build a new typed list (with a well-defined type) and copy the reflected list items to the typed list. This process can be quite expensive compared to the overall computation. Thus, this is often better not to use lists when arrays can be used instead (typically when you cannot know the size of the final list): Numpy array are significantly faster, more compact and functions using them can be compiled faster.
Additionally, Numba has no idea of was is the type Particle. Numba only supports Numpy built-in types by default. There is an experimental support for jitted classes but I advise you to work with basic array since it is generally faster and also more flexible as you can use Numpy vectorized functions on the target arrays as opposed to an object-based array (which are AFAIK inefficiently stored in Numpy arrays and slow).
Moreover, you should really avoid using global variables, especially the ones that are mutated. Global variables are slower to access in CPython and are generally seen as a bad software engineering practice. For Numba, they are considered as compile time constant so it can cause some surprising results if the variables are mutated at runtime.