I have a data frame, which has an embedded tree structure inside it, like this:
df1
Category Type Dependent-Category
0 1 ~ A
1 1 ~ B
2 1 ~ C
3 1 ~ 14
4 1 ~ D
5 1 P NaN
6 A ~ C
7 A ~ D
8 A ~ 3
9 A P NaN
10 B ~ D
11 B ~ C
12 B ~ 12
13 B P NaN
14 C ~ D
15 C ~ 9
16 C P NaN
17 D ~ 12
18 D ~ 3
19 D ~ 8
20 D P NaN
Category D rows are only made out of numerical categories, which is defined as.
D ~ 12
D ~ 3
D ~ 8
D P NaN
Category C rows are made out of a numerical Category and the previous Category D, which is defined as:
C ~ D
C ~ 9
C P NaN
Category B are made out of a numerical Category and the previous 2 Categories D and C, which is defined as:
B ~ D
B ~ C
B ~ 12
B P NaN
Category A are made out of a numerical Category and the previous 2 Categories C and D, which is defined as:
A ~ C
A ~ D
A ~ 3
A P NaN
Category 1 are made out of a numerical Category and the previous 4 Categories A,B,C and D, which is defined as: 1 ~ A 1 ~ B 1 ~ C 1 ~ 14 1 ~ D 1 P NaN
The aim is to get a final dataframe which has is a total concatenation of all the Categories - for example, when Category A is mentioned in the Dependent-Category column in df1, I need to replace that entire row with Category A.
The aim is to make the Dependent-Category column only consist of numbers (and NaNs):
CodePudding user response:
Possible solution can be to separate out different categories as dataframes and then merge them sequentially with original dataframe then apply conditional formatting.
Here is my solution doing the same:
d = {
"Category": [1, 1, 1, 1, "A", "A", "A", "B", "B"],
"Type": ["~", "~", "~", "P", "~", "~", "P", "~", "P"],
"D_C": ["B", "A", 14, "missing", 4, "B", "missing", 12, "missing"],
}
df = pd.DataFrame(d)
df_A = df.loc[df.Category == "A"]
df_B = df.loc[df.Category == "B"]
df1 = df.merge(df_A, left_on=df.D_C, right_on=df_A.Category, how="outer")
df1_A = pd.DataFrame()
df1_A["Category"] = np.where(
df1.D_C_x == df1.Category_y, df1.Category_y, df1.Category_x
)
df1_A["Type"] = np.where(df1.D_C_x == df1.Category_y, df1.Type_y, df1.Type_x)
df1_A["D_C"] = np.where(df1.D_C_x == df1.Category_y, df1.D_C_y, df1.D_C_x)
df2 = df1_A.merge(df_B, left_on=df1_A.D_C, right_on=df_B.Category, how="outer")
df_final = pd.DataFrame()
df_final["Category"] = np.where(
df2.D_C_x == df2.Category_y, df2.Category_y, df2.Category_x
)
df_final["Type"] = np.where(df2.D_C_x == df2.Category_y, df2.Type_y, df2.Type_x)
df_final["D_C"] = np.where(df2.D_C_x == df2.Category_y, df2.D_C_y, df2.D_C_x)
print(df_final)
Let me know if you have questions around it.
CodePudding user response:
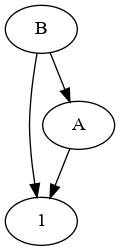
I would approach this using graph theory.
The advantage is that you do not need to worry about the depth of the mappings (e.g., B->A->C->D would be tricky, even worse if you have circular paths).
You have the following directed graph:
You can use networkx to generate your graph from the part of your DataFrame that has a dependent category, and to map the childrens automatically:
import networkx as nx
# split the DataFrame in twao parts
# df1 will contain the rows with a dependent category
# df2 will contain the rows with the values
# using here a regex to identify letter but any other logic can be used
m = df['Dependent-Category'].str.match('[a-zA-Z] ').fillna(False)
df1 = df[m]
df2 = df[~m]
# generating the graph
G = nx.from_pandas_edgelist(df1, create_using=nx.DiGraph,
source='Dependent-Category', target='Category')
# keep only nodes with children
nodes = set(filter(G.out_degree, G.nodes))
# {'A', 'B'}
# mapping nodes to children and self
d = {n: (nx.descendants(G, n)&nodes)|{n} for n in nodes}
# {'A': {'A'}, 'B': {'A', 'B'}}
# replacing the values in the DataFrame
df3 = (df1['Dependent-Category']
.map(d).explode()
.to_frame('Category')
.reset_index()
.merge(df2)
.set_index('index')
)
# merging the above df3 to df2
out = pd.concat([df2, df3]).sort_index(kind='stable')
output:
Category Type Dependent-Category
0 A ~ 4
0 A P NaN
0 B ~ 12
0 B P NaN
1 A ~ 4
1 A P NaN
2 1 ~ 14
3 1 P NaN
4 A ~ 4
5 A ~ 4
5 A P NaN
5 B ~ 12
5 B P NaN
6 A P NaN
7 B ~ 12
8 B P NaN