I have data as follows:
dat <- structure(list(rn = c("A", "B", "C",
"D", "E"), `[0,25)` = c("40 (replaced)",
"52 (replaced)", "5", "2", "5 (replaced)"), `[25,50)` = c("0 (replaced)",
"0 (replaced)", "0 (replaced)", "0 (replaced)", "0 (replaced)"), `[25,100)` = c("5",
"3", "38", "2", "1"), `[50,100)` = c("0 (replaced)", "0 (replaced)",
"0 (replaced)", "0 (replaced)", "0 (replaced)")), row.names = c(NA,
-5L), class = c("data.table", "data.frame"))
rn [0,25) [25,50) [25,100) [50,100)
1: A 40 (replaced) 0 (replaced) 5 0 (replaced)
2: B 52 (replaced) 0 (replaced) 3 0 (replaced)
3: C 5 0 (replaced) 38 0 (replaced)
4: D 2 0 (replaced) 2 0 (replaced)
5: E 5 (replaced) 0 (replaced) 1 0 (replaced)
I could simply get the numbers out as follows:
dat <- t(apply(dat, 1, extract_numeric))
dat <- as.data.frame(dat )
dat <- dat %>%
rowwise() %>%
summarise(V1 = V1, freq =list(c_across(-V1))) %>%
rowwise() %>%
mutate(freq = list(freq[which(freq > 0)]))
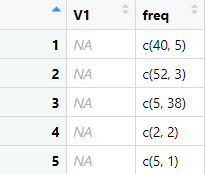
dat_out <- structure(list(V1 = c(NA_real_, NA_real_, NA_real_, NA_real_,
NA_real_), freq = list(c(40, 5), c(52, 3), c(5, 38), c(2, 2),
c(5, 1))), class = c("rowwise_df", "tbl_df", "tbl", "data.frame"
), row.names = c(NA, -5L), groups = structure(list(.rows = structure(list(
1L, 2L, 3L, 4L, 5L), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), row.names = c(NA, -5L), class = c("tbl_df",
"tbl", "data.frame")))
But how should I do it if I want to keep the text as well?
Desired output:
freq
c("40 (replaced)","5")
c("52 (replaced)","3")
c("5","38")
c("2","2")
c("5 (replaced)","1")
CodePudding user response:
It may be easier after reshaping to 'long' format with pivot_longer, filter out the rows having '0' values in the 'value' column with a regex match, then grouped by 'rn', summarise the 'value' elements in a list
library(dplyr)
library(tidyr)
library(stringr)
out <- dat %>%
pivot_longer(cols = -rn) %>%
filter(str_detect(value, '\\b0\\b', negate = TRUE)) %>%
group_by(rn) %>%
summarise(freq = list(value), .groups = 'drop')
-output
> out
# A tibble: 5 × 2
rn freq
<chr> <list>
1 A <chr [2]>
2 B <chr [2]>
3 C <chr [2]>
4 D <chr [2]>
5 E <chr [2]>
> out$freq
[[1]]
[1] "40 (replaced)" "5"
[[2]]
[1] "52 (replaced)" "3"
[[3]]
[1] "5" "38"
[[4]]
[1] "2" "2"
[[5]]
[1] "5 (replaced)" "1"
Or another option is to replace the column elements with 0 to NA, then unite to a single column, specifying the na.rm = TRUE and if needed split into a list with strsplit on the delimiter ,
dat %>%
mutate(across(-rn, ~ replace(.x,
str_detect(.x, '\\b0\\b'), NA_character_))) %>%
unite(freq, -rn, na.rm = TRUE, sep=",") %>%
mutate(freq = strsplit(freq, ","))
rn freq
<char> <list>
1: A 40 (replaced),5
2: B 52 (replaced),3
3: C 5,38
4: D 2,2
5: E 5 (replaced),1