I want to plot a density histogram in R with discrete data. Given that my dataset is very large I need to compute the densities prior plotting.
However, I find that using the stats::density function provides different results compared to ..density.. in ggplot2. Why is that? Also the ..density.. results from ggplot2 are in line with what is expected whereas stats::density are not.
See reproducible example below.
Many Thanks
library(tidyverse)
library(patchwork)
df <- data.frame(A = round(rnorm(1000)),
B = round(rnorm(1000)),
C = round(rnorm(1000))) %>%
pivot_longer(cols = everything(), names_to = "group")
dens_df <- df %>%
group_by(group) %>%
summarise(dens = list(density(value, from = -3, to = 6, n = length(-3:6)))) %>% #compute density and nest into list
mutate(density.x = map(dens, ~.x[["x"]]), #extract x values
density.y = map(dens, ~.x[["y"]])) %>% #extract y values
select(-dens) %>%
unnest(cols = c(density.x, density.y))
plot_dens <- dens_df %>%
ggplot()
aes(x = density.x, y = density.y) %>%
geom_col()
scale_x_continuous(breaks = seq(-3,10,1))
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1), geom = "point", col = "red")
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1), geom = "point", size = 2, col = "red")
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1), geom = "line", col = "red")
facet_wrap(~group)
labs(title = "using stats::density")
plot_geom <- df %>%
ggplot()
aes(x = value, y = ..density..) %>%
geom_histogram(
binwidth = 1,
col = "white")
scale_x_continuous(breaks = seq(-3,10,1))
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1), geom = "point", col = "red")
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1), geom = "point", size = 2, col = "red")
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1), geom = "line", col = "red")
facet_wrap(~group)
labs(title = "using ggplot2 ..density..")
plot_dens plot_geom
CodePudding user response:
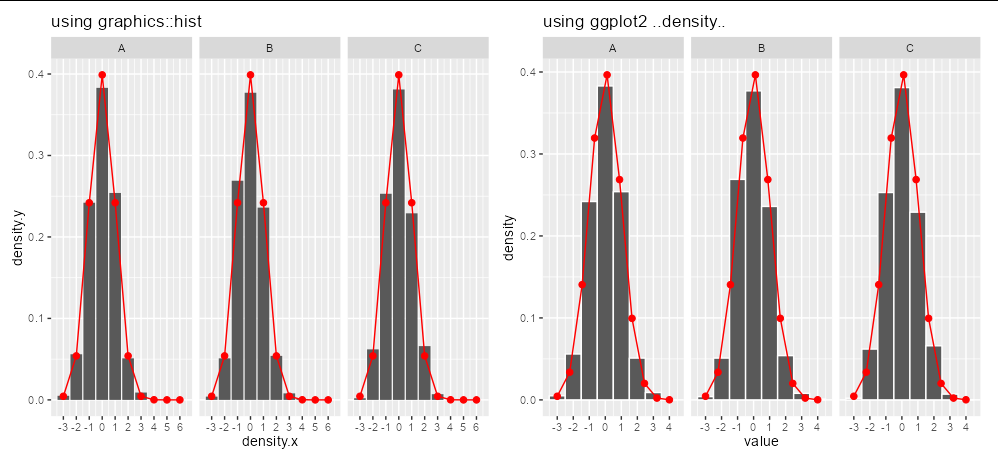
The simple answer is that stats::density isn't the correct function to use on discrete data. stats::density uses a smoothing kernel and will provide a curve representing the continuous density between -3 and 6, even if only sampled at 10 points. This is not the same as the densities of discrete data, which is just the frequency of each bin divided by the number of observations. This is what geom_histogram is drawing.
The computationally efficient way to get discrete densities is to use hist or even just table(x)/length(x).
Here's an example using precomputed densities from hist
dens_df <- df %>%
group_by(group) %>%
summarise(dens = list(hist(value, breaks = seq(-3.5, 6.5), plot = FALSE))) %>%
mutate(density.x = map(dens, ~.x[["mids"]]), #extract x values
density.y = map(dens, ~.x[["density"]])) %>% #extract y values
select(-dens) %>%
unnest(cols = c(density.x, density.y))
We can see this leads to the same plot as geom_histogram, using your own plotting code but with the altered dens_df
plot_dens <- dens_df %>%
ggplot()
aes(x = density.x, y = density.y) %>%
geom_col()
scale_x_continuous(breaks = seq(-3,10,1))
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1),
geom = "point", col = "red")
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1),
geom = "point", size = 2, col = "red")
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1),
geom = "line", col = "red")
facet_wrap(~group)
labs(title = "using graphics::hist")
plot_geom <- df %>%
ggplot()
aes(x = value, y = ..density..) %>%
geom_histogram(
binwidth = 1,
col = "white")
scale_x_continuous(breaks = seq(-3,10,1))
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1),
geom = "point", col = "red")
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1),
geom = "point", size = 2, col = "red")
stat_function(fun = dnorm, n = 10, args = list(mean = 0, sd = 1),
geom = "line", col = "red")
facet_wrap(~group)
labs(title = "using ggplot2 ..density..")
plot_dens plot_geom