I am producing a heatmap via cooccur package but I am getting overlapping species names. How to solve it?
The plot was produced by the following codes:
M <- cooccur(mat = M, type = "spp_site", thresh = T, spp_names = TRUE, prob = "hyper")
plot(M, plotrand = TRUE)
CodePudding user response:
Unfortunately, the author didn't use defined themes inside the function. In this post 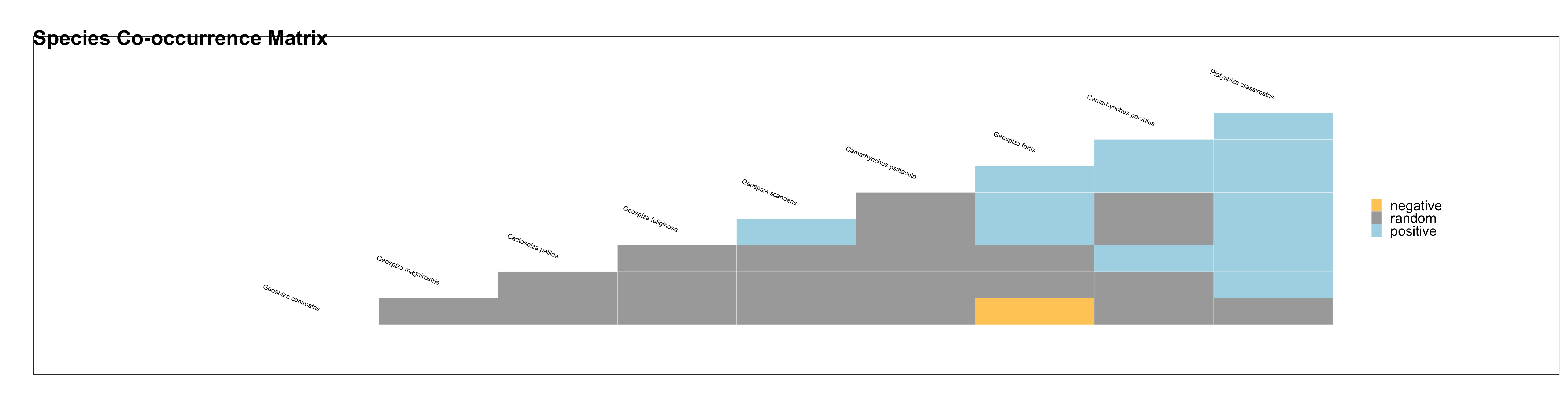
I would suggest playing around with the theme options from ggplot. Here is a good post which can help you for sure: Changing font size and direction of axes text in ggplot2
CodePudding user response:
Before you start fiddling with font sizes, you should check whether this is just an artificial problem caused by R incorrectly guessing the device size. I have noticed that this regularly occurs when a plot is saved to PDF with dev.copy and the following workaround always solved the problem for me:
- Resize the plotting window on screen.
- Redraw the plot by calling the plot command again.
