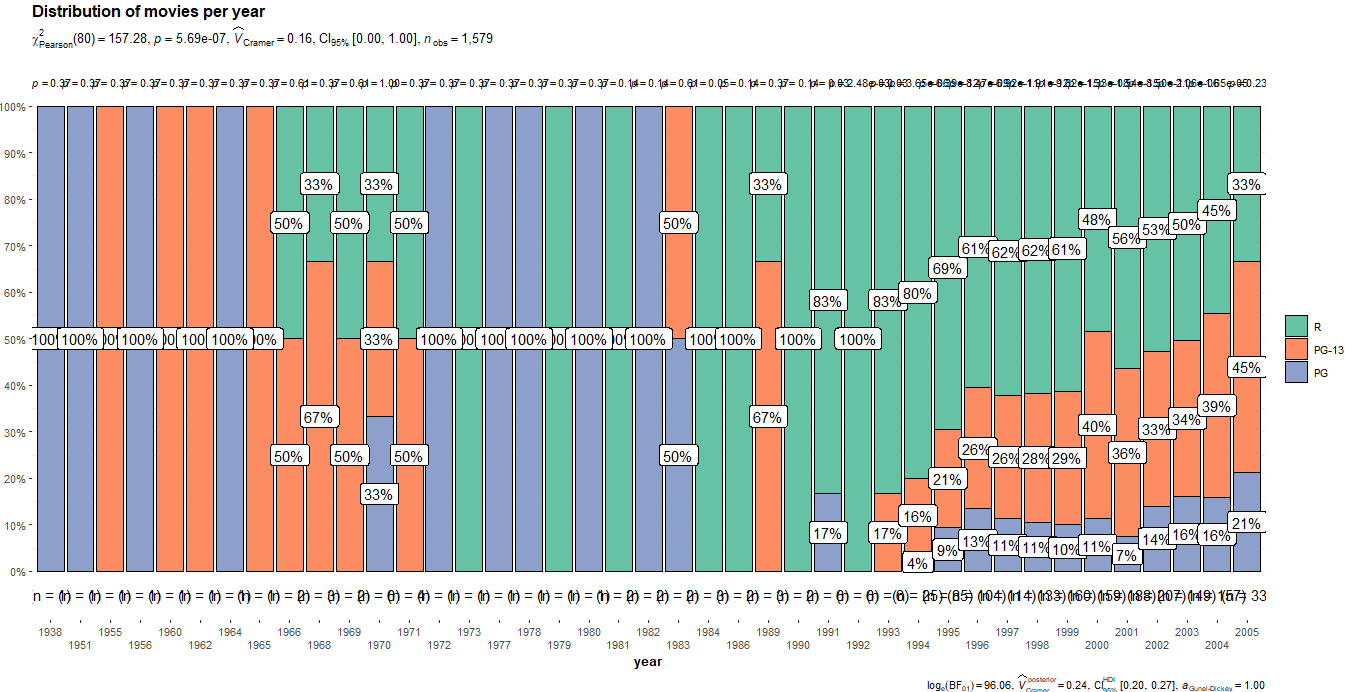
I am working on a plot using ggstatsplot R package. After completing the plot I have an issue. This is the code for my plot:
library(ggstatsplot)
#Data
data("movies_long")
movies_long
#Plot
Myplot <- ggbarstats(
data = movies_long,
x = mpaa,
y = year,
title = "Distribution of movies per year",
xlab = "year",
legend.title = "",
ggplot.component = list(ggplot2::scale_x_discrete(guide = ggplot2::guide_axis(n.dodge = 2))),
palette = "Set2"
)
The output is next:
The plot is very useful but it has two problems. First is the overlap of p-values at top of bars. Second is the size n which also has overlap, how could I improve the p-values labels at the top of bars and also the size n at bottom?
Is there any way to take these p-values and translate in p<0.001, p<0.1 using confidence levels?
Many thanks for your help. In the extreme case, is there any way to know how to compute the p-values (which test is being applied) at each bar, so that I can replicate it and create a similar plot using ggplot2?
CodePudding user response:
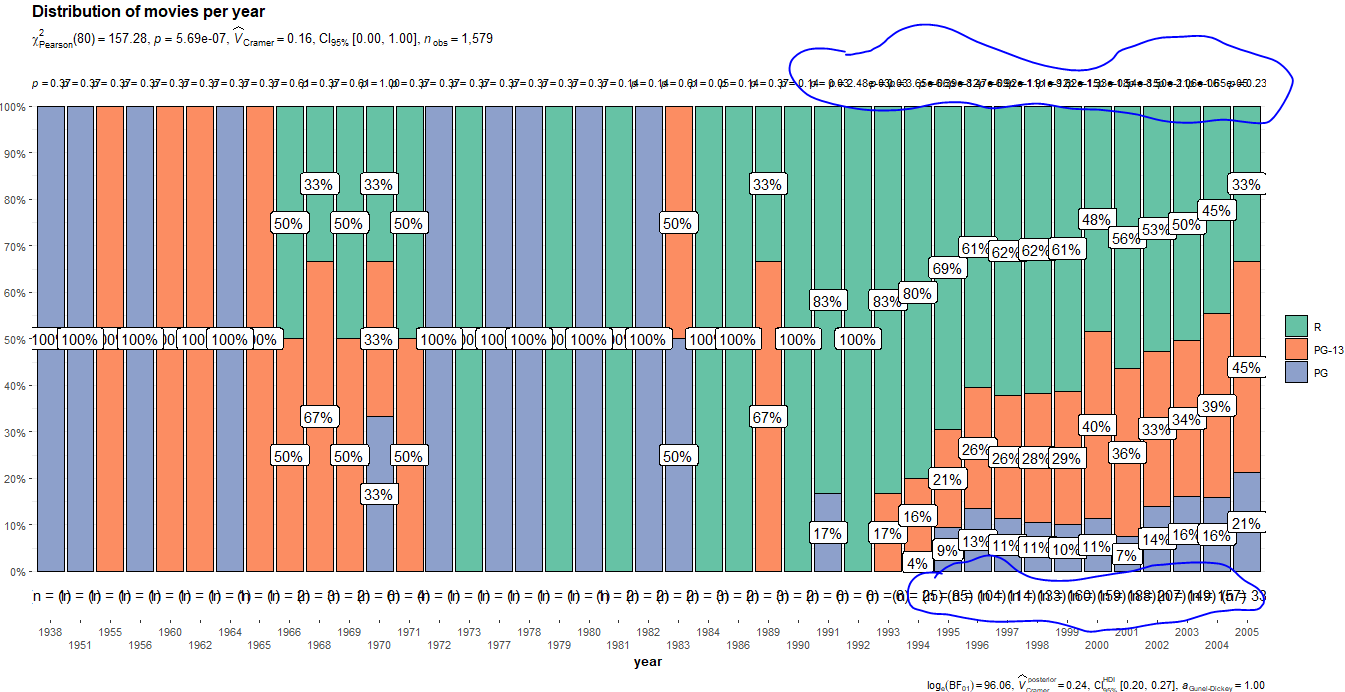
I challenge your statement that this plot is very useful. Apart from the overlapping labels, there are statistical issues including:
- The plot almost completely obscures the fact that it summarizes 1 movie per year for most years before 1991.
- The analysis computes lots of p-values without any correction for multiple hypothesis testing, which is bad practice.
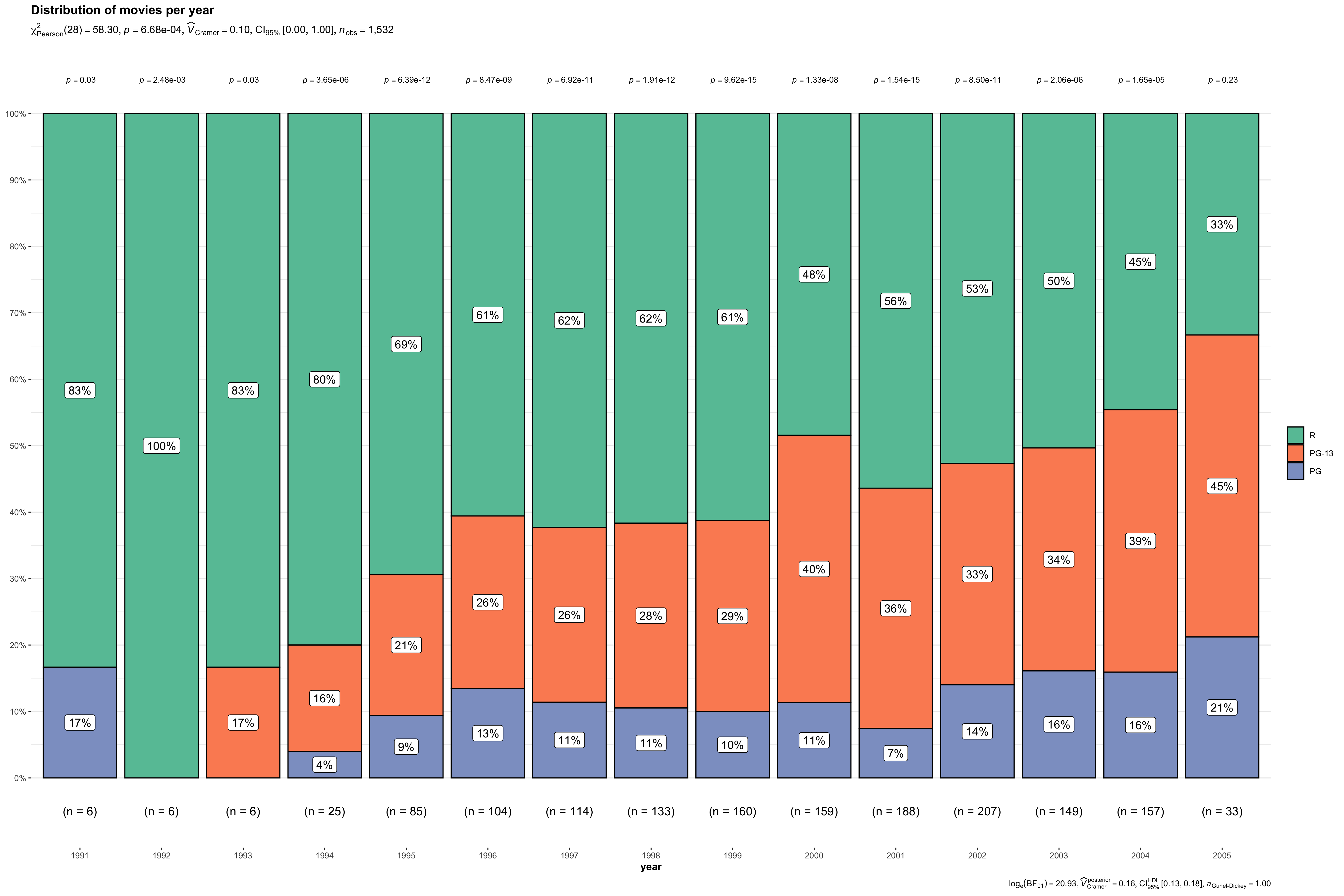
Here is the same plot restricted to movies released between 1991 and 2005. It's better, though still peppered with overabundance of uncorrected p-values.
Note: I've made the plot large enough to accommodate all those labels. Now the text is a bit small but with some trial and error you can get the plot height & width "right".
As for the p-values on top of each bar... There is a handy way to extract the data visualized in a ggplot:
p$data
#> # A tibble: 41 × 5
#> year mpaa counts perc .label
#> <fct> <fct> <int> <dbl> <chr>
#> 1 1991 R 5 83.3 83%
#> 2 1992 R 6 100 100%
#> 3 1993 R 5 83.3 83%
#> 4 1994 R 20 80 80%
#> 5 1995 R 59 69.4 69%
#> 6 1996 R 63 60.6 61%
#> 7 1997 R 71 62.3 62%
#> 8 1998 R 82 61.7 62%
#> 9 1999 R 98 61.3 61%
#> 10 2000 R 77 48.4 48%
#> # … with 31 more rows
Now that we've confirmed that each bar is a simple summary of the number of movies by audience rating (R, PG, PG-13), we can quickly check that the p-value comes from a chi-squared test on the counts by rating:
p$data %>%
filter(
year == 2000
)
#> # A tibble: 3 × 5
#> year mpaa counts perc .label
#> <fct> <fct> <int> <dbl> <chr>
#> 1 2000 R 77 48.4 48%
#> 2 2000 PG-13 64 40.3 40%
#> 3 2000 PG 18 11.3 11%
chisq.test(c(77, 64, 18))
#>
#> Chi-squared test for given probabilities
#>
#> data: c(77, 64, 18)
#> X-squared = 36.264, df = 2, p-value = 1.335e-08