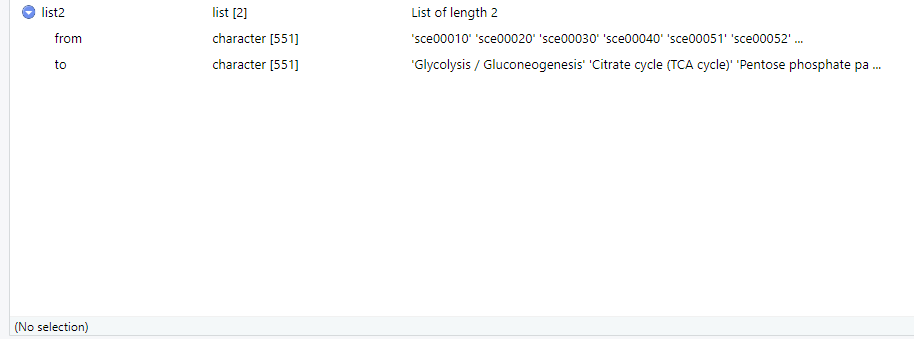
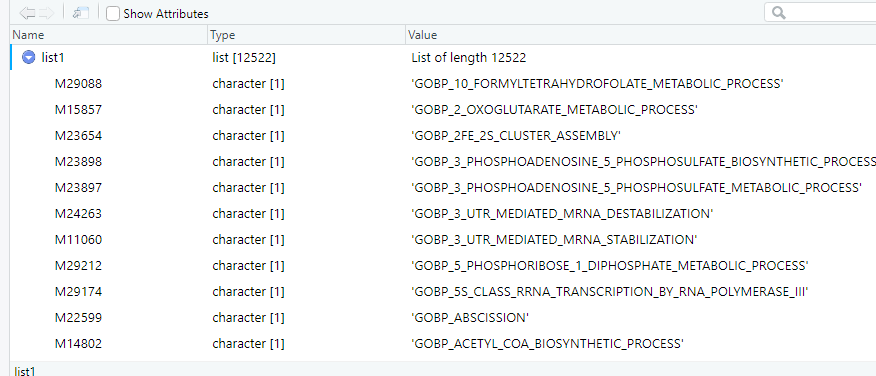
I would like to convert the format of my list2 to be like list1, where those letters in "from" go to Names column and whatever that is in "to" go to Value column like in list1. In list1, from and to values are associated. For example, "sce00010" goes with "Glycolysis/Glucogneogeneis".
Is there any way to do so? Tnx!
Structure of list2:
structure(list(from = c("sce00010", "sce00020", "sce00030", "sce00040",
"sce00051", "sce00052"), to = c("Glycolysis / Gluconeogenesis",
"Citrate cycle (TCA cycle)", "Pentose phosphate pathway", "Pentose and glucuronate interconversions",
"Fructose and mannose metabolism", "Galactose metabolism")), row.names = c(NA,
6L), class = "data.frame")
from to
1 sce00010 Glycolysis / Gluconeogenesis
2 sce00020 Citrate cycle (TCA cycle)
3 sce00030 Pentose phosphate pathway
4 sce00040 Pentose and glucuronate interconversions
5 sce00051 Fructose and mannose metabolism
6 sce00052 Galactose metabolism
structure of list1:
c(sce00010= "Glycolysis / Gluconeogenesis",
sce00020 = "Citrate cycle (TCA cycle)", sce00030= "Pentose phosphate pathway",
sce00040 = "Pentose and glucuronate interconversions",
sce00051= "Fructose and mannose metabolism",
sce00052= "Galactose metabolism")
sce00010
"Glycolysis / Gluconeogenesis"
sce00020
"Citrate cycle (TCA cycle)"
sce00030
"Pentose phosphate pathway"
sce00040
"Pentose and glucuronate interconversions"
sce00051
"Fructose and mannose metabolism"
sce00052
"Galactose metabolism"
CodePudding user response:
list1 <- list2$to
names(list1) <- list2$from
list1
#> sce00010
#> "Glycolysis / Gluconeogenesis"
#> sce00020
#> "Citrate cycle (TCA cycle)"
#> sce00030
#> "Pentose phosphate pathway"
#> sce00040
#> "Pentose and glucuronate interconversions"
#> sce00051
#> "Fructose and mannose metabolism"
#> sce00052
#> "Galactose metabolism"
Created on 2022-06-19 by the reprex package (v2.0.1)