I'm trying to visualize a grouped boxplot, where the data for group 1 is from one dataset, and for another group is from another dataset.
I wish to plot four groups of box plots (each group having two box plots, each one from each of the two datasets) based on the condition that in the data frame the last column contains four specific names. So a box plot for every different name.
I thought of rbind() and combining the two datasets, but the column names are different, so that shall not work.
The dataset1 is:
structure(list(ZNF8 = c(6.535976, 5.617546, 5.520076, 6.173408,
5.636716, 5.600262, 5.220575, 4.743357, 4.901029, 4.925283, 5.838014,
4.596382, 4.914675, 5.377216, 5.555028, 5.314098, 5.297864, 5.750704,
5.488065, 4.999103, 5.992091, 5.642773, 5.07518, 5.314475, 5.900937,
5.559608, 4.522685, 5.265154, 5.324593, 5.354034, 5.7289, 5.057386,
5.06977, 5.597334, 5.153986, 5.108475, 5.570181, 4.899142, 5.406266,
5.113253, 4.960403, 5.435193, 6.867495, 4.856415, 5.111619, 5.449276,
5.198133, 5.388347, 5.282403, 4.632593, 4.829716, 4.811047, 5.32258,
4.610439, 5.18732, 5.64755, 5.482928, 5.119356, 5.444986, 5.477327,
4.885934, 5.349004, 5.66351, 4.303683, 4.592721, 5.422557, 5.087755,
4.716865, 4.669027, 5.340175, 4.782326, 5.649563, 5.132957, 5.683762,
5.932028, 7.316127, 6.146593, 5.72388, 5.966438, 4.792467, 5.720676,
5.148644, 5.73848, 6.212466, 6.308748, 5.379114, 5.272329, 5.439909,
5.586977, 5.09346, 5.576321, 6.207303, 5.592295, 4.593619, 4.889057,
5.293568, 4.586061, 5.428235, 5.8397, 5.96753, 5.801161, 5.987631,
6.203965, 4.839773, 4.715386, 4.636048, 5.860811, 6.033741, 4.733141,
5.138581, 5.114714, 5.466906, 4.908799, 5.060534, 5.255573, 4.830971,
5.263964, 5.525848, 5.220203, 4.957148, 4.740335, 4.5796, 4.594491,
5.352114, 5.157587, 5.436527, 4.91852, 5.08691, 5.023482, 5.534429,
5.051983, 5.253279, 5.040294, 5.216557, 4.901129, 5.324232, 4.74494,
5.136233, 5.024926, 5.33796, 4.793476, 4.707207, 5.811963, 5.502805,
5.038211, 4.890697, 4.606382), ZNF804A = c(6.51568, 6.439824,
4.177717, 5.722987, 7.93036, 8.825149, 6.117847, 5.202625, 6.943736,
6.10238, 5.395202, 4.005275, 5.449567, 5.250869, 5.375079, 6.279801,
4.846173, 4.312459, 4.968536, 7.3861, 8.261597, 7.374785, 6.239196,
5.368542, 5.101474, 5.054613, 5.31017, 4.846913, 5.216192, 5.737364,
5.338054, 4.207094, 6.386935, 4.983061, 7.574587, 4.413107, 5.352411,
6.507691, 5.199225, 5.900246, 5.338985, 4.046878, 8.342389, 4.816628,
5.722269, 4.635663, 4.8078, 4.223784, 5.340559, 5.434575, 4.493881,
4.84106, 6.921161, 4.690488, 6.626063, 4.473161, 4.489687, 4.323735,
4.360158, 4.583291, 6.180486, 4.410217, 6.529253, 4.332168, 5.635954,
4.46047, 5.444109, 4.409367, 4.607851, 4.415356, 3.9169, 4.028548,
4.286836, 4.498551, 5.981919, 4.136005, 4.24244, 4.089674, 4.29059,
3.859174, 4.062029, 4.326703, 4.236188, 4.27184, 3.95776, 4.491866,
4.209833, 4.202512, 4.158955, 4.174552, 5.370753, 4.933963, 3.982912,
4.162634, 4.101793, 4.332177, 4.263031, 5.336449, 4.02567, 4.168412,
4.131943, 4.263566, 4.107345, 4.111515, 4.241965, 4.483141, 4.170102,
4.146675, 4.479603, 4.473624, 4.667704, 4.019978, 4.190319, 4.442643,
4.456675, 5.083862, 4.649441, 4.57102, 4.110476, 4.423792, 4.461519,
4.309157, 3.784853, 4.249824, 4.127431, 4.136897, 4.779541, 4.261828,
4.150655, 4.605232, 4.489233, 4.284094, 5.285193, 4.413362, 4.528692,
4.108301, 5.526067, 5.096504, 4.480856, 4.271599, 5.242742, 4.213557,
4.74649, 4.027772, 4.640365, 4.344877, 4.077101), ttcluster_dataset_1.Subtype = c("Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Proneural (Cluster 1)",
"Proneural (Cluster 1)", "Proneural (Cluster 1)", "Neural (Cluster 1)",
"Neural (Cluster 1)", "Neural (Cluster 1)", "Neural (Cluster 1)",
"Neural (Cluster 1)", "Neural (Cluster 1)", "Neural (Cluster 1)",
"Neural (Cluster 1)", "Neural (Cluster 1)", "Neural (Cluster 1)",
"Neural (Cluster 1)", "Neural (Cluster 1)", "Neural (Cluster 1)",
"Neural (Cluster 1)", "Neural (Cluster 1)", "Neural (Cluster 1)",
"Neural (Cluster 1)", "Neural (Cluster 1)", "Neural (Cluster 1)",
"Neural (Cluster 1)", "Neural (Cluster 1)", "Neural (Cluster 1)",
"Neural (Cluster 1)", "Neural (Cluster 1)", "Neural (Cluster 1)",
"Neural (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Classical (Cluster 1)", "Classical (Cluster 1)", "Classical (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)",
"Mesenchymal (Cluster 1)", "Mesenchymal (Cluster 1)")), row.names = c("TCGA.02.0014.01",
"TCGA.02.0026.01", "TCGA.02.0048.01", "TCGA.02.0069.01", "TCGA.02.0074.01",
"TCGA.02.0080.01", "TCGA.02.0084.01", "TCGA.02.0087.01", "TCGA.02.0104.01",
"TCGA.02.0114.01", "TCGA.02.0281.01", "TCGA.02.0321.01", "TCGA.02.0325.01",
"TCGA.02.0338.01", "TCGA.02.0339.01", "TCGA.02.0432.01", "TCGA.02.0439.01",
"TCGA.02.0440.01", "TCGA.02.0446.01", "TCGA.06.0128.01", "TCGA.06.0129.01",
"TCGA.06.0146.01", "TCGA.06.0156.01", "TCGA.06.0166.01", "TCGA.06.0174.01",
"TCGA.06.0177.01", "TCGA.06.0238.01", "TCGA.06.0241.01", "TCGA.06.0410.01",
"TCGA.06.0413.01", "TCGA.06.0414.01", "TCGA.06.0646.01", "TCGA.06.0648.01",
"TCGA.08.0245.01", "TCGA.08.0344.01", "TCGA.08.0347.01", "TCGA.08.0348.01",
"TCGA.08.0350.01", "TCGA.08.0353.01", "TCGA.08.0359.01", "TCGA.08.0385.01",
"TCGA.08.0517.01", "TCGA.08.0524.01", "TCGA.12.0616.01", "TCGA.12.0618.01",
"TCGA.02.0089.01", "TCGA.02.0113.01", "TCGA.02.0115.01", "TCGA.02.0451.01",
"TCGA.06.0132.01", "TCGA.06.0133.01", "TCGA.06.0138.01", "TCGA.06.0160.01",
"TCGA.06.0162.01", "TCGA.06.0167.01", "TCGA.06.0171.01", "TCGA.06.0173.01",
"TCGA.06.0179.01", "TCGA.06.0182.01", "TCGA.06.0185.01", "TCGA.06.0195.01",
"TCGA.06.0208.01", "TCGA.06.0214.01", "TCGA.06.0219.01", "TCGA.06.0221.01",
"TCGA.06.0237.01", "TCGA.06.0240.01", "TCGA.08.0349.01", "TCGA.08.0380.01",
"TCGA.08.0386.01", "TCGA.08.0520.01", "TCGA.02.0016.01", "TCGA.02.0023.01",
"TCGA.02.0070.01", "TCGA.02.0102.01", "TCGA.02.0260.01", "TCGA.02.0269.01",
"TCGA.02.0285.01", "TCGA.02.0289.01", "TCGA.02.0290.01", "TCGA.02.0317.01",
"TCGA.02.0333.01", "TCGA.02.0422.01", "TCGA.02.0430.01", "TCGA.06.0125.01",
"TCGA.06.0126.01", "TCGA.06.0137.01", "TCGA.06.0145.01", "TCGA.06.0148.01",
"TCGA.06.0187.01", "TCGA.06.0211.01", "TCGA.06.0402.01", "TCGA.08.0246.01",
"TCGA.08.0354.01", "TCGA.08.0355.01", "TCGA.08.0357.01", "TCGA.08.0358.01",
"TCGA.08.0375.01", "TCGA.08.0511.01", "TCGA.08.0514.01", "TCGA.08.0518.01",
"TCGA.08.0529.01", "TCGA.08.0531.01", "TCGA.02.0004.01", "TCGA.02.0039.01",
"TCGA.02.0059.01", "TCGA.02.0064.01", "TCGA.02.0075.01", "TCGA.02.0079.01",
"TCGA.02.0085.01", "TCGA.02.0086.01", "TCGA.02.0099.01", "TCGA.02.0106.01",
"TCGA.02.0107.01", "TCGA.02.0111.01", "TCGA.02.0326.01", "TCGA.02.0337.01",
"TCGA.06.0122.01", "TCGA.06.0124.01", "TCGA.06.0143.01", "TCGA.06.0147.01",
"TCGA.06.0149.01", "TCGA.06.0152.01", "TCGA.06.0154.01", "TCGA.06.0164.01",
"TCGA.06.0175.01", "TCGA.06.0176.01", "TCGA.06.0184.01", "TCGA.06.0189.01",
"TCGA.06.0190.01", "TCGA.06.0194.01", "TCGA.06.0197.01", "TCGA.06.0210.01",
"TCGA.06.0397.01", "TCGA.06.0409.01", "TCGA.06.0412.01", "TCGA.06.0644.01",
"TCGA.06.0645.01", "TCGA.08.0346.01", "TCGA.08.0352.01", "TCGA.08.0360.01",
"TCGA.08.0390.01", "TCGA.08.0509.01", "TCGA.08.0510.01", "TCGA.08.0512.01",
"TCGA.12.0619.01", "TCGA.12.0620.01"), class = "data.frame")
The dataset2 is:
structure(list(ZNF8 = c(5.119095, 4.760636, 5.416521, 5.363732,
4.495227, 4.902833, 6.109712, 4.667864, 5.015435, 5.181961, 5.834061,
4.864606, 5.081433, 5.220504, 4.869951, 4.930496, 4.52962, 4.493444,
4.526999, 4.635502, 5.040377, 4.765426, 4.493035, 5.218385),
ZNF804A = c(4.192456, 4.246963, 6.287769, 4.228797, 4.226257,
4.474805, 4.336341, 4.362907, 4.182984, 4.173398, 4.151008,
4.37511, 4.066581, 4.050416, 4.283787, 4.534188, 4.24613,
4.056275, 4.55178, 4.29397, 5.215219, 4.295077, 4.434866,
5.129624), ttcluster_dataset_2.Subtype = c("Proneural (Cluster 2)",
"Proneural (Cluster 2)", "Proneural (Cluster 2)", "Proneural (Cluster 2)",
"Proneural (Cluster 2)", "Proneural (Cluster 2)", "Proneural (Cluster 2)",
"Proneural (Cluster 2)", "Classical (Cluster 2)", "Classical (Cluster 2)",
"Classical (Cluster 2)", "Classical (Cluster 2)", "Classical (Cluster 2)",
"Classical (Cluster 2)", "Mesenchymal (Cluster 2)", "Mesenchymal (Cluster 2)",
"Mesenchymal (Cluster 2)", "Mesenchymal (Cluster 2)", "Mesenchymal (Cluster 2)",
"Mesenchymal (Cluster 2)", "Mesenchymal (Cluster 2)", "Mesenchymal (Cluster 2)",
"Mesenchymal (Cluster 2)", "Mesenchymal (Cluster 2)")), row.names = c("TCGA.02.0003.01",
"TCGA.02.0010.01", "TCGA.02.0011.01", "TCGA.02.0024.01", "TCGA.02.0028.01",
"TCGA.02.0046.01", "TCGA.02.0047.01", "TCGA.02.0060.01", "TCGA.02.0007.01",
"TCGA.02.0009.01", "TCGA.02.0021.01", "TCGA.02.0027.01", "TCGA.02.0038.01",
"TCGA.02.0043.01", "TCGA.02.0025.01", "TCGA.02.0033.01", "TCGA.02.0034.01",
"TCGA.02.0051.01", "TCGA.02.0054.01", "TCGA.02.0057.01", "TCGA.06.0130.01",
"TCGA.06.0139.01", "TCGA.08.0392.01", "TCGA.08.0522.01"), class = "data.frame")
My attempt has been:
par(mfrow=(1,1))
ttcluster_dataset_11 %>%
tidyr::pivot_longer(cols = -ttcluster_dataset_1.Subtype) %>%
ggplot()
geom_boxplot(aes(
x =ttcluster_dataset_1.Subtype,
y = value,
color=ttcluster_dataset_1.Subtype,
fill= ttcluster_dataset_1.Subtype))
labs(x = "Phenotypes",
y = "Gene Expression Value")
scale_fill_brewer(palette="RdBu") guides(fill=guide_legend(title="Phenotypes"),color=guide_legend(title="Phenotypes")) ggtitle("Cluster 1 \nGene Expression across Phenotypes")
ttcluster_dataset_22 %>%
tidyr::pivot_longer(cols = -ttcluster_dataset_2.Subtype) %>%
ggplot()
geom_boxplot(aes(
x =ttcluster_dataset_2.Subtype,
y = value,
color=ttcluster_dataset_2.Subtype,
fill= ttcluster_dataset_2.Subtype))
labs(x = "Phenotypes",
y = "Gene Expression Value")
scale_fill_brewer(palette="RdBu") guides(fill=guide_legend(title="Phenotypes"),color=guide_legend(title="Phenotypes")) ggtitle("Cluster 2 \nGene Expression across Phenotypes")
All suggestions shall be apprecited.
CodePudding user response:
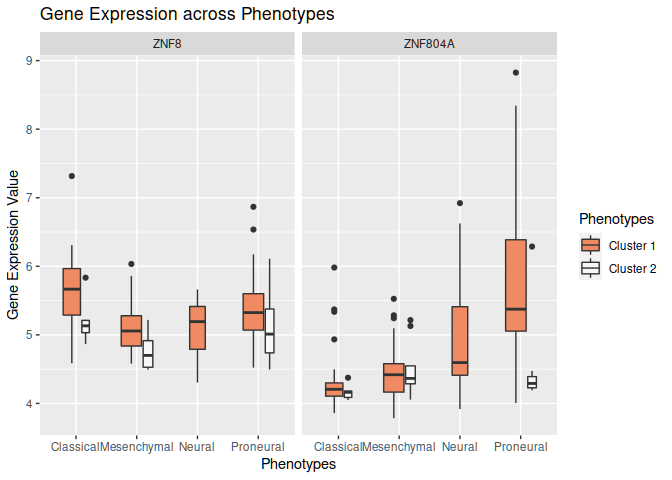
One option would be to rename the columns, add an additional column to designate the clusters and do an rbind. You may also consider to use facet_wrap to split by the two genes:
library(dplyr)
library(ggplot2)
dataset1 <- dataset1 %>%
tidyr::pivot_longer(cols = -ttcluster_dataset_1.Subtype) %>%
rename(Subtype=ttcluster_dataset_1.Subtype) %>%
mutate(Cluster="Cluster 1", Subtype=gsub(" .*", "", Subtype))
dataset2 <- dataset2 %>%
tidyr::pivot_longer(cols = -ttcluster_dataset_2.Subtype) %>%
rename(Subtype=ttcluster_dataset_2.Subtype) %>%
mutate(Cluster="Cluster 2", Subtype=gsub(" .*", "", Subtype))
rbind(dataset1, dataset2) %>% ggplot(aes(x=Subtype, y=value, fill=Cluster))
geom_boxplot(varwidth = TRUE)
facet_wrap(~name)
labs(x = "Phenotypes",
y = "Gene Expression Value")
scale_fill_brewer(palette="RdBu")
guides(fill=guide_legend(title="Phenotypes"),color=guide_legend(title="Phenotypes"))
ggtitle("Gene Expression across Phenotypes")