I'm having trouble getting the plots I need from a mixed effects regression, and wonder if I could get some help.
I would like to get a dot plot of days on X axis, and OR to have a reaction time slower than 300ms on the Y axis, to see the effect over time.
Using the sleepstudy data from lme4 as simple sample data. I coerced "day" into a "factor" so that the model would give an OR for each day
df <- data.frame(sleepstudy)
# Generate a categorical value for reaction time to be slower than 300ms
df <- df %>%
mutate(slow = ifelse(Reaction >= 300, 1,0), Days = as.factor(Days))
# Mixed effects model with subject as random effect
library(lme4)
m <- glmer(slow ~ Days (1 | Subject), data=df, family=binomial)
library(sjPlot)
plot_model(m)
2 issues:
The above code gives me a forest plot, which is kind of what I want, except i would like it flipped on the side with the OR on the Y axis and days running along the X axis.
My real data has 8 predictors, and I would like to only plot the effect for the predictor "Days" - however once I convert it to a factor, the argument "terms" needs me to spell out every day individual , i.e.
plot_model(m, terms = c("Days1", "Days2", "Days3" etc))
The alternative is to rm.terms, but that means I need to type out 7 terms to remove. Is there a more efficient way for it to just plot all levels along "Days"?
CodePudding user response:
Perhaps something like this helps as a starting point?
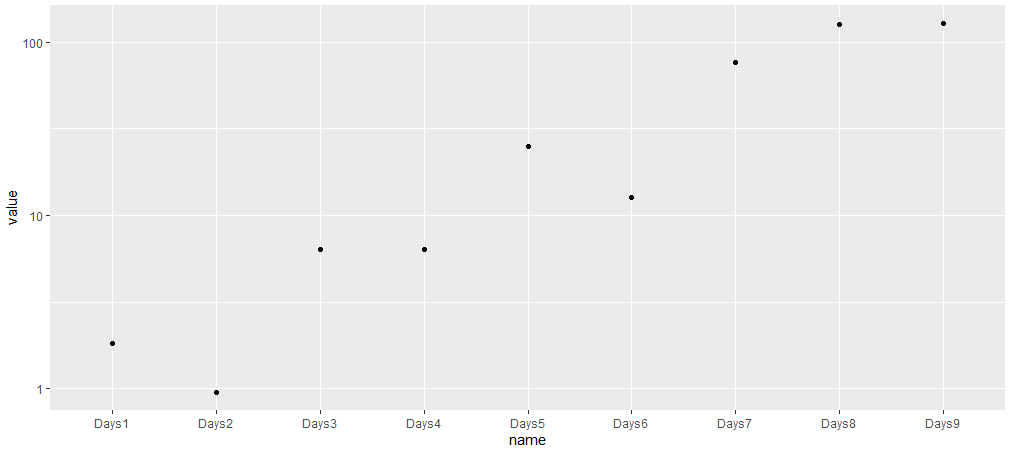
library(tidyverse)
m %>%
fixef() %>%
enframe() %>%
filter(!str_detect(name, "Intercept")) %>%
mutate(value = exp(value)) %>%
ggplot(aes(name, value))
geom_point()
scale_y_log10()
Some further comments:
- If you have more days, you probably need to make sure that the factor levels have the correct order; i.e. you need to avoid that Day10 comes before Day2 (which will happen with the default lexicographic ordering).
- These are only the fixed effect point estimates. You could also modify the code to include confidence intervals.
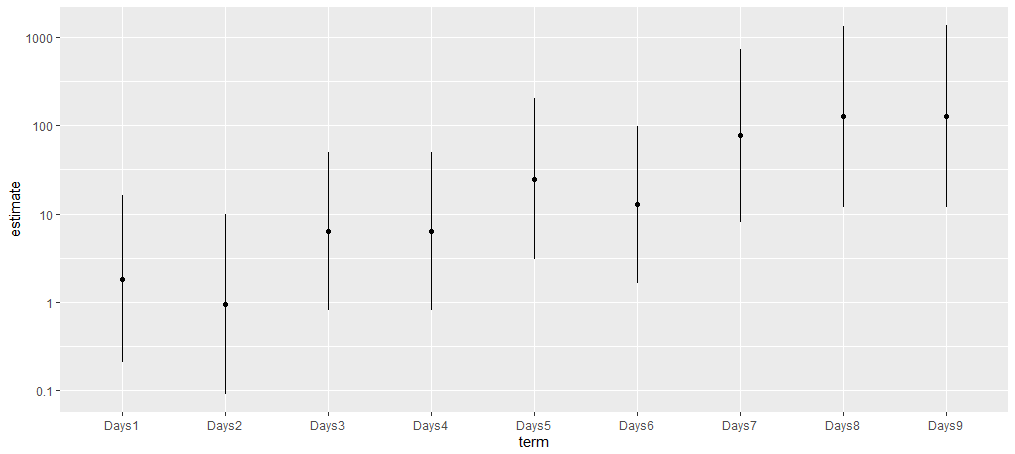
To include confidence bands for the estimates you can use the fantastic broom.mixed extension to get a tidy table of estimates.
library(broom.mixed)
m %>%
tidy(effects = "fixed", conf.int = TRUE) %>%
filter(!str_detect(term, "Intercept")) %>%
mutate(across(c(estimate, starts_with("conf")), exp)) %>%
ggplot(aes(term, estimate))
geom_point()
geom_linerange(aes(ymin = conf.low, ymax = conf.high))
scale_y_log10(labels = function(x) sprintf("%g", x))