I have a large dataset of mineral nitrogen values from different plots which includes some missing data were on some dates we could not take samples. it is known that mineral N values in soil change linearly between samplings.
for the sake of simplification I have created a data frame that has 10 plots with 4 dates (with different distances between them) with missing data in one of the dates:
df <- data.frame(plot= c(1,2,3,4,5,6,7,8,9,10),
date = c("2020-10-01", "2020-10-01","2020-10-01","2020-10-01","2020-10-01","2020-10-01","2020-10-01","2020-10-01","2020-10-01","2020-10-01",
"2020-10-08", "2020-10-08","2020-10-08","2020-10-08","2020-10-08","2020-10-08","2020-10-08","2020-10-08","2020-10-08","2020-10-08",
"2020-10-29","2020-10-29","2020-10-29","2020-10-29","2020-10-29","2020-10-29","2020-10-29","2020-10-29","2020-10-29","2020-10-29",
"2020-11-05","2020-11-05","2020-11-05","2020-11-05","2020-11-05","2020-11-05","2020-11-05","2020-11-05","2020-11-05","2020-11-05"),
Nmin = c(100, 120, 50, 60, 70, 80, 100, 70, 30, 50, 90, 130, 60, 60, 60, 90, 105, 60, 25, 40, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, 50, 170, 100, 60, 20, 130, 125, 20, 5, 0))
df$date <- as.Date(df$date, format="%d.%m.%Y")
df$Nmin <- as.numeric(df$Nmin)
is there a function that can calculate the missing values of Nmin plot-wise and takes in concideration the time between samplings (date)?
CodePudding user response:
Using approx.
df <- transform(df, flag=ifelse(is.na(Nmin), 1, 0)) ## set flag for sake of identification
res <- by(df, df$plot, \(x) {
transform(x, Nmin=approx(x$date, x$Nmin, x$date)$y)
}) |> unsplit(df$plot)
res
# plot date Nmin flag
# 1 1 2020-10-01 100 0
# 2 2 2020-10-01 120 0
# 3 3 2020-10-01 50 0
# 4 4 2020-10-01 60 0
# 5 5 2020-10-01 70 0
# 6 6 2020-10-01 80 0
# 7 7 2020-10-01 100 0
# 8 8 2020-10-01 70 0
# 9 9 2020-10-01 30 0
# 10 10 2020-10-01 50 0
# 11 1 2020-10-08 90 0
# 12 2 2020-10-08 130 0
# 13 3 2020-10-08 60 0
# 14 4 2020-10-08 60 0
# 15 5 2020-10-08 60 0
# 16 6 2020-10-08 90 0
# 17 7 2020-10-08 105 0
# 18 8 2020-10-08 60 0
# 19 9 2020-10-08 25 0
# 20 10 2020-10-08 40 0
# 21 1 2020-10-29 60 1
# 22 2 2020-10-29 160 1
# 23 3 2020-10-29 90 1
# 24 4 2020-10-29 60 1
# 25 5 2020-10-29 30 1
# 26 6 2020-10-29 120 1
# 27 7 2020-10-29 120 1
# 28 8 2020-10-29 30 1
# 29 9 2020-10-29 10 1
# 30 10 2020-10-29 10 1
# 31 1 2020-11-05 50 0
# 32 2 2020-11-05 170 0
# 33 3 2020-11-05 100 0
# 34 4 2020-11-05 60 0
# 35 5 2020-11-05 20 0
# 36 6 2020-11-05 130 0
# 37 7 2020-11-05 125 0
# 38 8 2020-11-05 20 0
# 39 9 2020-11-05 5 0
# 40 10 2020-11-05 0 0
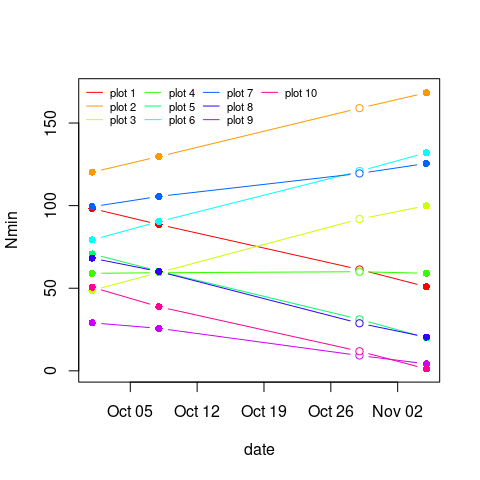
Let's take a look at the plot.
clr <- rainbow(10)
with(res, plot(Nmin ~ date, type='n'))
by(res, res$plot, \(x) with(x, points(jitter(Nmin) ~ date, type='b', pch=ifelse(flag == 1, 21, 16), col=clr[plot], bg='white')))
legend('topleft', legend=paste('plot', 1:10), lty=1, col=clr, ncol=4, bty='n', cex=.7)
Note: For non-linear inter/extrapolation, see this answer.
Data:
df <- structure(list(plot = c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 1, 2,
3, 4, 5, 6, 7, 8, 9, 10, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 1, 2,
3, 4, 5, 6, 7, 8, 9, 10), date = structure(c(18536, 18536, 18536,
18536, 18536, 18536, 18536, 18536, 18536, 18536, 18543, 18543,
18543, 18543, 18543, 18543, 18543, 18543, 18543, 18543, 18564,
18564, 18564, 18564, 18564, 18564, 18564, 18564, 18564, 18564,
18571, 18571, 18571, 18571, 18571, 18571, 18571, 18571, 18571,
18571), class = "Date"), Nmin = c(100, 120, 50, 60, 70, 80, 100,
70, 30, 50, 90, 130, 60, 60, 60, 90, 105, 60, 25, 40, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, 50, 170, 100, 60, 20, 130, 125,
20, 5, 0), flag = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0)), class = "data.frame", row.names = c(NA, -40L
))