I am calculating the weighted average of a series of datetimes (and must be doing it wrong, since I can't explain the following):
import pandas as pd
import numpy as np
foo = pd.DataFrame({'date': ['2022-06-01', '2022-06-16'],
'value': [1000, 10000]})
foo['date'] = pd.to_datetime(foo['date'])
bar = np.average(foo['date'].view(dtype='float64'), weights=foo['value'])
print(np.array(bar).view(dtype='datetime64[ns]'))
returns
2022-06-14T15:16:21.818181818, which is expected.
Changing the month to July:
foo = pd.DataFrame({'date': ['2022-07-01', '2022-07-16'],
'value': [1000, 10000]})
foo['date'] = pd.to_datetime(foo['date'])
bar = np.average(foo['date'].view(dtype='float64'), weights=foo['value'])
print(np.array(bar).view(dtype='datetime64[ns]'))
returns 2022-07-14T23:59:53.766924660,
when the expected result is
2022-07-14T15:16:21.818181818.
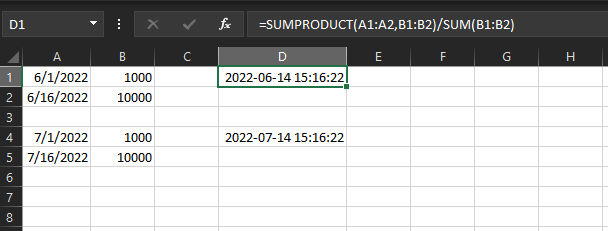
The expected result, as calculated in Excel:
What am I overlooking?
EDIT: Additional Detail
- My real dataset is much larger and I'd like to use numpy if possible.
foo['date']can be assumed to be dates with no time component, but the weighted average will have a time component.
CodePudding user response:
I strongly suspect this is a resolution/rounding issue.
I'm assuming that for averaging dates those are converted to timestamps - and then the result is converted back to a datetime object. But pandas works in nanoseconds, so the timestamp values, multiplied by 1000 and 10000 respectively, exceed 2**52 - i.e. exceed the mantissa capability of 64-bit floats.
On the contrary Excel works in milliseconds, so no problems here; Python's datetime.datetime works in microseconds, so still no problems:
dt01 = datetime(2022,7,1)
dt16 = datetime(2022,7,16)
datetime.fromtimestamp((dt01.timestamp()*1000 dt16.timestamp()*10000)/11000)
datetime.datetime(2022, 7, 14, 15, 16, 21, 818182)
So if you need to use numpy/pandas I suppose your best option is to convert dates to timedeltas from a "starting" date (i.e. define a "custom epoch") and compute the weighted average of those values.
CodePudding user response:
first of all I don't think the problem is in your code, I think pandas has a problem.
the problem is that when you use the view command it translates it to a very small number (e-198) so I believe it accidently lose resolution.
I found a solution (I think it works but it gave an 3 hour difference from your answer):
from datetime import datetime
import pandas as pd
import numpy as np
foo = pd.DataFrame({'date': ['2022-07-01', '2022-07-16'],
'value': [1000, 10000]})
foo['date'] = pd.to_datetime(foo['date'])
# bar = np.average([x.timestamp() for x in foo['date']], weights=foo['value'])
bar = np.average(foo['date'].apply(datetime.timestamp), weights=foo['value']) # vectorized
print(datetime.fromtimestamp(bar))