I am working on building a classs. But I realized that one part is extremly slow. I believe it is this part below, because when I call it, it takes couple of minutes which is too long. How can the speed of the function be improved? Not sure how the code can be improved to increase speed.
def ability_metrics(self, TPR,FPR,TNR,FNR):
TPR_p = chisquare(list(np.array(list(TPR.values()))*100))[1]
FPR_p = chisquare(list(np.array(list(FPR.values()))*100))[1]
TNR_p = chisquare(list(np.array(list(TNR.values()))*100))[1]
FNR_p = chisquare(list(np.array(list(FNR.values()))*100))[1]
if TPR_p <= 0.01:
print("*** Reject H0: Significant True Positive Disparity with p=",TPR_p)
elif TPR_p <= 0.05:
print("** Reject H0: Significant True Positive Disparity with p=",TPR_p)
elif TPR_p <= 0.1:
print("* Reject H0: Significant True Positive Disparity with p=",TPR_p)
else:
print("Accept H0: True Positive Disparity Not Detected. p=",TPR_p)
if FPR_p <= 0.01:
print("*** Reject H0: Significant False Positive Disparity with p=",FPR_p)
elif FPR_p <= 0.05:
print("** Reject H0: Significant False Positive Disparity with p=",FPR_p)
elif FPR_p <= 0.1:
print("* Reject H0: Significant False Positive Disparity with p=",FPR_p)
else:
print("Accept H0: False Positive Disparity Not Detected. p=",FPR_p)
if TNR_p <= 0.01:
print("*** Reject H0: Significant True Negative Disparity with p=",TNR_p)
elif TNR_p <= 0.05:
print("** Reject H0: Significant True Negative Disparity with p=",TNR_p)
elif TNR_p <= 0.1:
print("* Reject H0: Significant True Negative Disparity with p=",TNR_p)
else:
print("Accept H0: True Negative Disparity Not Detected. p=",TNR_p)
if FNR_p <= 0.01:
print("*** Reject H0: Significant False Negative Disparity with p=",FNR_p)
elif FNR_p <= 0.05:
print("** Reject H0: Significant False Negative Disparity with p=",FNR_p)
elif FNR_p <= 0.1:
print("* Reject H0: Significant False Negative Disparity with p=",FNR_p)
else:
print("Accept H0: False Negative Disparity Not Detected. p=",FNR_p)
def predictive(self, labels,sens_df):
precision_dic = {}
for i in labels:
precision_dic[labels[i]] = precision_score(sens_df[labels[i]]['t'],sens_df[labels[i]]['p'])
fig = go.Figure([go.Bar(x=list(labels.values()), y=list(precision_dic.values()))])
pred_p = chisquare(list(np.array(list(precision_dic.values()))*100))[1]
return(precision_dic,fig,pred_p)
There is a second part which also include couple of if else statements.
def identify_bias(self, sensitive,labels):
predictions = self.model.predict(self.X_test)
cont_table,sens_df,rep_fig,rep_p = self.representation(sensitive,labels,predictions)
print("REPRESENTATION")
rep_fig.show()
print(cont_table,'\n')
if rep_p <= 0.01:
print("*** Reject H0: Significant Relation Between",sensitive,"and Target with p=",rep_p)
elif rep_p <= 0.05:
print("** Reject H0: Significant Relation Between",sensitive,"and Target with p=",rep_p)
elif rep_p <= 0.1:
print("* Reject H0: Significant Relation Between",sensitive,"and Target with p=",rep_p)
else:
print("Accept H0: No Significant Relation Between",sensitive,"and Target Detected. p=",rep_p)
TPR, FPR, TNR, FNR = self.ability(sens_df,labels)
print("\n\nABILITY")
self.ability_plots(labels,TPR,FPR,TNR,FNR)
self.ability_metrics(TPR,FPR,TNR,FNR)
precision_dic, pred_fig, pred_p = self.predictive(labels,sens_df)
print("\n\nPREDICTIVE")
pred_fig.show()
if pred_p <= 0.01:
print("*** Reject H0: Significant Predictive Disparity with p=",pred_p)
elif pred_p <= 0.05:
print("** Reject H0: Significant Predictive Disparity with p=",pred_p)
elif pred_p <= 0.1:
print("* Reject H0: Significant Predictive Disparity with p=",pred_p)
else:
print("Accept H0: No Significant Predictive Disparity. p=",pred_p)
CodePudding user response:
If your chisquare is scipy.stats.chisquare, then it takes numpy arrays so you can just use np.fromiter(your_dict.values(), dtype=float) * 100 (or dtype=int as required) as your argument instead of converting it to a list, then to an array, then to a list again.
Even if your function doesn't take numpy arrays and absolutely must have a list for some reason, you could consider iterating directly over your_dict.values() and multiplying the elements of that iterator in a list comprehension. [i * 100 for i in your_dict.values()], or using np.ndarray.tolist() to convert the numpy array to a list.
Out of interest, I wrote some code to time the different approaches.
- Direct conversion from
dicttondarray, multiply by 100 dicttolist, then tondarray, multiply by 100, then tolistdicttondarray, multiply by 100, thenlistdicttondarray, multiply by 100, thennp.tolistdicttolistusing list comprehension
import timeit
import numpy as np
from matplotlib import pyplot as plt
# This is a function decorator that I can use to set the func.plot_args and func.plot_kwargs
# attributes, which I use later in the plot function
def plot_args(*args, **kwargs):
def _iargs(func):
func.plot_args = args or list()
func.plot_kwargs = kwargs or dict()
return func
return _iargs
@plot_args(label="dict->array")
def f0(x):
return np.fromiter(x.values(), dtype=int) * 100
@plot_args(label="dict->list->array->list")
def f1(x):
return list(np.array(list(x.values())) * 100)
@plot_args(label="dict->array->list")
def f2(x):
return list(np.fromiter(x.values(), dtype=int) * 100)
@plot_args(label="dict->array->np.tolist")
def f3(x):
return (np.fromiter(x.values(), dtype=int) * 100).tolist()
@plot_args(label="dict->list")
def f4(x):
return [i * 100 for i in x.values()]
funcs = [f0, f1, f2, f3, f4]
times = []
N = [int(10**i) for i in (0, 1, 2, 3, 4, 5, 6, 7)]
num = 50
for n in N:
mult = 1 if n < 1000 else 10
x = {i: i for i in range(n)}
times.append([])
for f in funcs:
t = timeit.timeit('f(x)', globals=globals(), number=num//mult)
times[-1].append(t*mult)
print(f"n={n}; f={f.__name__}; t={t}")
times = np.array(times)
# fig, (ax, iax) = plt.subplots(2, 1, sharex=True, figsize=(6.4, 10))
fig, ax = plt.subplots(1, 1)
iax = ax.inset_axes([0.08, 0.5, 0.40, 0.47])
for fi, fn in enumerate(funcs):
ln, = ax.plot(N, times[:, fi], *fn.plot_args, **fn.plot_kwargs)
iax.plot(N, times[:, fi], *fn.plot_args, **fn.plot_kwargs)
ax.legend(loc="lower right")
ax.set_xscale('log')
ax.set_yscale('log')
ax.set_ylabel(f"Time for {num} function calls (s)")
ax.set_xlabel("Dict size")
ax.grid(color="lightgray")
iax.set_xlim([N[-3], ax.get_xlim()[1]*0.6])
iax.set_ylim([times[-3:, :].min()/1.1, times[-3:].max()*1.1])
iax.set_xscale('log')
iax.grid(color="lightgray", alpha=0.5)
ax.indicate_inset_zoom(iax, edgecolor='black')
plt.tight_layout()
plt.show()
This gives the following plot:
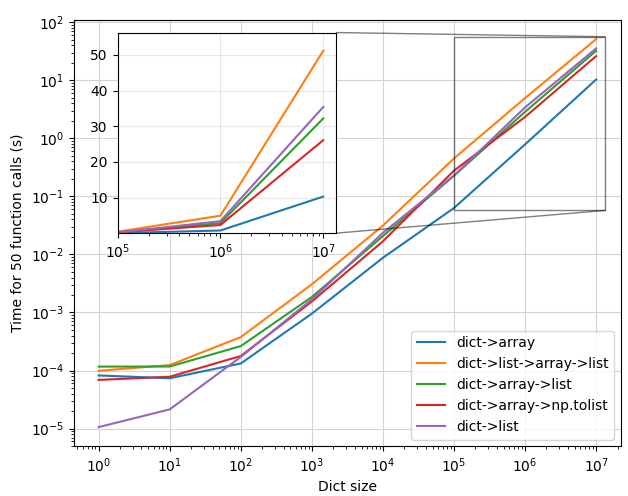
Some observations:
- At small values of dictionary size ( < 100 ), the
dict->listapproach (f4) is fastest by far. Next aredict->array(f0) anddict->array->np.tolist(f3). Finally, we havedict->array->list(f2), anddict->list->array->list(f1). This makes sense -- it takes time to convert everything to a numpy array, so if your dictionary is small enough, just multiply it in python and get it over with.f1is predictably on the slower side, since it involves the most conversions, and therefore the most work. Interestingly,f2is similar in performance tof1, which would indicate that converting a numpy array to a list usinglist()is a bottleneck. - At larger input sizes ( > 10,000 ),
f4becomes slower thanf0, and slowly gets worse than the other approaches that use numpy. Converting the numpy array to a list is consistently faster usingnp.ndarray.tolist()than usinglist(...) - The inset shows the zoomed-in plot at the largest values. The inset y axis has a linear scale to better demonstrate the differences in each method at scale, and clearly
f1is by far the slowest, andf0by far the fastest.
To conclude:
- If your dicts are small, use a list comprehension.
- If your dicts are large, don't convert to a list, use a numpy array
- If your dicts are large and you must convert to a list, avoid
list(...). Usenp.fromiterto convert the dict directly to a numpy array, andnp.ndarray.tolist()to convert the numpy array to a list.
