Data
Here is my data:
work <- structure(list(Mins_Sleep = c(300L, 540L, 540L, 480L, 480L, 480L,
480L, 420L, 300L, 240L, 480L, 300L, 420L, 360L, 390L, 405L, 420L,
360L, 420L, 350L, 420L, 450L, 445L, 480L, 300L, 400L, 310L, 390L,
350L, 450L, 390L, 390L, 510L, 452L, 310L, 360L, 500L, 360L, 420L,
420L, 382L, 430L, 393L, 240L, 400L, 480L, 450L, 450L, 359L, 420L,
361L, 360L, 480L, 570L, 340L, 450L, 420L, 180L, 510L, 420L, 425L,
407L, 360L, 510L, 360L, 368L, 410L, 360L, 510L, 436L, 291L, 420L,
240L, 300L, 420L, 420L, 424L, 520L, 240L, 390L, 480L, 300L, 480L,
390L, 300L, 360L, 420L, 360L, 480L, 330L, 375L, 390L, 458L, 517L,
435L, 482L, 300L, 589L, 440L, 391L, 420L, 545L, 574L, 529L, 420L,
579L, 317L, 532L, 506L, 346L, 559L, 402L, 444L, 450L, 405L, 480L,
438L, 477L, 480L, 300L, 420L, 240L, 390L, 420L, 420L, 300L, 360L,
360L, 360L, 585L, 300L, 420L, 420L, 304L, 420L, 420L, 300L, 467L,
230L, 460L, 400L, 513L, 498L, 240L, 360L, 320L, 420L, 224L, 396L,
360L, 300L, 360L, 360L, 480L, 412L, 822L, 300L, 300L, 416L, 360L,
420L, 430L, 370L, 360L, 420L, 420L, 420L, 173L, 360L, 319L, 420L,
360L, 415L, 540L, 370L, 504L, 480L, 525L, 300L, 480L, 360L, 360L,
360L, 420L, 420L, 570L, 420L, 360L, 420L, 420L, 450L, 360L, 450L,
480L, 420L, 360L, 420L, 420L, 420L, 360L, 420L, 420L, 420L, 480L,
300L, 360L, 420L, 485L, 420L, 462L, 331L, 483L, 494L, 360L, 514L,
349L, 408L, 480L, 480L, 480L, 447L, 363L, 442L, 441L, 407L, 522L,
300L, 300L, 405L, 405L, 471L, 90L, 449L, 420L, 450L, 447L, 499L,
401L, 429L, 430L, 381L, 407L, 440L, 388L, 414L, 500L, 468L, 287L,
420L, 423L, 420L, 355L, 477L, 495L, 455L, 300L, 427L, 471L, 361L,
300L, 360L, 480L, 367L, 357L, 443L, 438L, 390L, 240L, 393L, 350L,
427L, 480L, 400L, 706L, 359L, 257L, 481L, 60L, 400L, 465L, 434L,
460L, 435L, 519L, 365L), Time_Wake = c(500L, 715L, 600L, 600L,
700L, 600L, 700L, 500L, 500L, 500L, 500L, 700L, 645L, 700L, 630L,
645L, 700L, 600L, 700L, 550L, 700L, 730L, 725L, 800L, 600L, 640L,
600L, 730L, 650L, 830L, 630L, 630L, 830L, 722L, 641L, 800L, 720L,
700L, 700L, 700L, 622L, 710L, 632L, 400L, 640L, 700L, 730L, 830L,
659L, 800L, 701L, 700L, 900L, 930L, 650L, 930L, 700L, 300L, 830L,
800L, 705L, 647L, 800L, 830L, 830L, 838L, 650L, 500L, 830L, 800L,
321L, 700L, 400L, 400L, 700L, 600L, 604L, 700L, 730L, 700L, 700L,
500L, 700L, 630L, 500L, 600L, 700L, 600L, 830L, 600L, 500L, 600L,
738L, 758L, 645L, 702L, 500L, 849L, 656L, 831L, 700L, 805L, 834L,
849L, 407L, 739L, 717L, 852L, 826L, 446L, 919L, 842L, 754L, 900L,
845L, 900L, 848L, 757L, 927L, 500L, 700L, 430L, 430L, 600L, 700L,
300L, 600L, 600L, 400L, 945L, 500L, 700L, 700L, 504L, 700L, 700L,
400L, 747L, 200L, 740L, 441L, 833L, 815L, 400L, 600L, 600L, 700L,
344L, 636L, 600L, 300L, 600L, 600L, 700L, 822L, 360L, 600L, 945L,
656L, 400L, 700L, 744L, 710L, 600L, 700L, 700L, 700L, 253L, 600L,
819L, 700L, 600L, 655L, 835L, 848L, 654L, 630L, 745L, 300L, 730L,
700L, 700L, 700L, 700L, 200L, 700L, 500L, 500L, 700L, 700L, 730L,
700L, 830L, 825L, 700L, 600L, 700L, 700L, 700L, 700L, 700L, 700L,
700L, 700L, 300L, 500L, 700L, 705L, 700L, 723L, 531L, 841L, 845L,
744L, 742L, 830L, 648L, 630L, 645L, 634L, 727L, 603L, 648L, 721L,
647L, 842L, 750L, 650L, 645L, 645L, 751L, 130L, 729L, 830L, 730L,
727L, 709L, 641L, 709L, 710L, 621L, 747L, 720L, 628L, 654L, 633L,
548L, 428L, 700L, 733L, 700L, 556L, 757L, 815L, 735L, 500L, 707L,
751L, 601L, 500L, 600L, 800L, 607L, 557L, 723L, 718L, 630L, 400L,
633L, 550L, 607L, 621L, 640L, 636L, 559L, 417L, 701L, 100L, 640L,
629L, 614L, 545L, 615L, 550L, 755L)), class = "data.frame", row.names = c(NA,
-285L))
Problem
I consistently see this kind of plot, but I never have found how to make it in R. When I google "scatterplot with residuals" or anything to that effect, I have problems finding this:
The best I know how to do so far is to make a standalone regression plot in ggplot:
library(tidyverse)
ggplot(work,
aes(x=Time_Wake,
y=Mins_Sleep))
geom_point(alpha=.4)
geom_smooth(method = "lm",
se=F,
color = "purple")
labs(title="An Attempt at Regression Plotting",
x="Onset Wake Time",
y="Minutes of Sleep")
However, I still don't know how to actually make the lines that represent the distance between raw values and the line of fit. How do I make this in R? I feel like this has to be something easy and I'm just not finding it.
CodePudding user response:
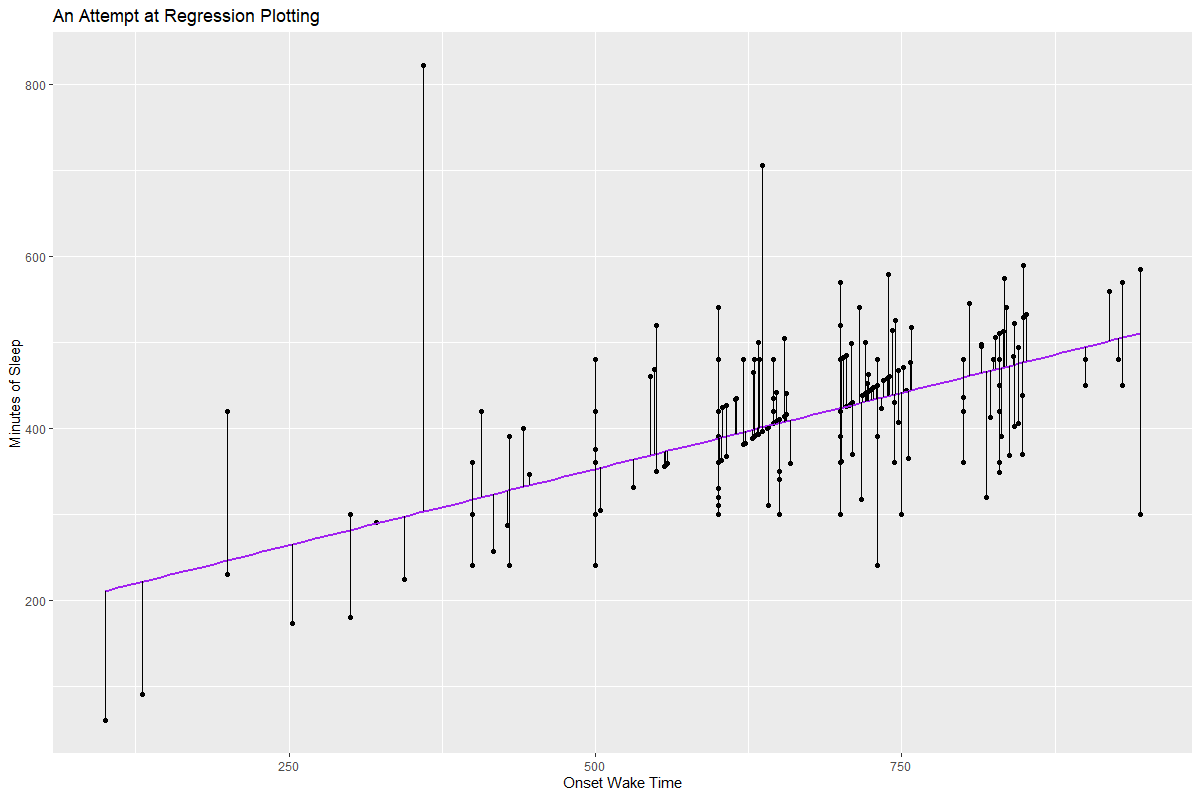
I would fit the lm to the data, then use broom::augment() to obtain the fitted values. You can then supply those to geom_segment.
library(ggplot2)
library(magrittr) # for pipes or load tidyverse
library(broom)
work %>%
lm(Mins_Sleep ~ Time_Wake, data = .) %>%
augment() %>%
ggplot(aes(Time_Wake, Mins_Sleep))
geom_point()
geom_smooth(method = "lm", se = FALSE, color = "purple")
geom_segment(aes(xend = Time_Wake, yend = .fitted))
labs(title = "An Attempt at Regression Plotting",
x = "Onset Wake Time",
y = "Minutes of Sleep")
Result:
This is what augment generates:
work %>%
lm(Mins_Sleep ~ Time_Wake, data = .) %>%
augment()
Result:
A tibble: 285 × 8
Mins_Sleep Time_Wake .fitted .resid .hat .sigma .cooksd .std.resid
<int> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 300 500 353. -52.5 0.00774 68.5 0.00232 -0.771
2 540 715 429. 111. 0.00408 68.2 0.00543 1.63
3 540 600 388. 152. 0.00407 67.9 0.0101 2.23
4 480 600 388. 92.0 0.00407 68.3 0.00371 1.35
5 480 700 423. 56.5 0.00382 68.5 0.00131 0.828
6 480 600 388. 92.0 0.00407 68.3 0.00371 1.35
7 480 700 423. 56.5 0.00382 68.5 0.00131 0.828
8 420 500 353. 67.5 0.00774 68.4 0.00382 0.990
9 300 500 353. -52.5 0.00774 68.5 0.00232 -0.771
10 240 500 353. -113. 0.00774 68.2 0.0106 -1.65
# … with 275 more rows