in the current graph the size of the nodes is given by the "count" of the arcs (e.g. node a going to a and b will be larger than the other two nodes that have no arcs to other nodes). the excel file is composed as follows:
- column 0 (date[0]) = value of the oriented arc
- column 1 (data[1]) = node a (node from where the oriented arc starts)
- column 2 (data[2]) = node b (node from where the directed arc arrives)
Code:
import networkx as nx
import matplotlib.pyplot as plt
import xlrd
file = '/a/example.xlsx'
G= nx.DiGraph()
nations = []
book = xlrd.open_workbook(file)
sheet=book.sheet_by_index(0)
for row in range(sheet.nrows):
data= sheet.row_slice(row)
Orig = data[1].value
Bene = data[2].value
nations.append((Orig, Bene))
pos = nx.planar_layout(G)
betCent = nx.betweenness_centrality(G, normalized=True, endpoints=True)
node_colors = [20000.0*G.degree(v) for v in G]
node_sizes = [v*7000 for v in betCent.values()]
plt.figure(figsize=(25,20))
nx.draw_networkx(G, pos=pos, with_labels=True,
node_color = node_colors,
node_size = node_sizes)
plt.axis('off')
G.add_edges_from(nations)
nx.draw(G,with_labels= True, verticalalignment='bottom')
plt.show()
Here the sample dataset:
df = pd.DataFrame({
'weight':['50'] * 4 ['500'] * 5 ['20'] * 3 ['100'],
'node a':['pippo', 'pippo', 'pippo', 'pluto', 'pippo', 'pippo',
'pluto', 'topolino', 'qua', 'minnie', 'winnie', 'pippo', 'qua'],
'node b':['pluto', 'qua', 'minnie', 'pluto', 'winnie', 'pippo',
'pluto', 'topolino', 'qua', 'qua', 'pippo', 'qua', 'qua'],
})
CodePudding user response:
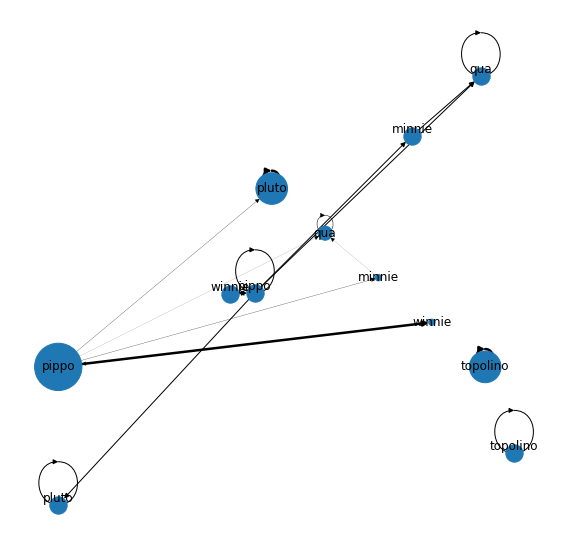
Below I show, how to calculate the sum of edge weights of outgoing edges and scale the nodes accordingly:
import networkx as nx
import matplotlib.pyplot as plt
import pandas as pd
df = pd.DataFrame({
'weight':['50'] * 4 ['500'] * 5 ['20'] * 3 ['100'],
'node a':['pippo', 'pippo', 'pippo', 'pluto', 'pippo', 'pippo',
'pluto', 'topolino', 'qua', 'minnie', 'winnie', 'pippo', 'qua'],
'node b':['pluto', 'qua', 'minnie', 'pluto', 'winnie', 'pippo',
'pluto', 'topolino', 'qua', 'qua', 'pippo', 'qua', 'qua'],
})
G= nx.DiGraph()
nations = []
for index, row in df.iterrows():
Orig = row[1]
Bene = row[2]
weight = float(row[0])
#add the wight of each edge as an attribute
nations.append((Orig, Bene, {'weight':weight}))
#generate Graph
G.add_edges_from(nations)
#iterate over nodes and sum the weight of their outgoing edges, save on graph as node attribute
nodes_weight = {}
for n in G.nodes():
nodes_weight[n]={'node_weight':sum([d['weight'] for o,b,d in G.out_edges(n, data=True)])}
nx.set_node_attributes(G, nodes_weight)
#draw graph
pos = nx.planar_layout(G)
#calculate node size from attribute
node_sizes = [v*2 for v in list(nx.get_node_attributes(G, 'node_weight').values())]
#similar if you want to change edge sizes
edge_sizes = [v/200 for v in list(nx.get_edge_attributes(G, 'weight').values())]
plt.figure(figsize=(10,10))
nx.draw_networkx(G, pos=pos, with_labels=True,
width = edge_sizes,
node_size = node_sizes)
plt.axis('off')
nx.draw(G,with_labels= True, verticalalignment='bottom')
plt.show()
Output: