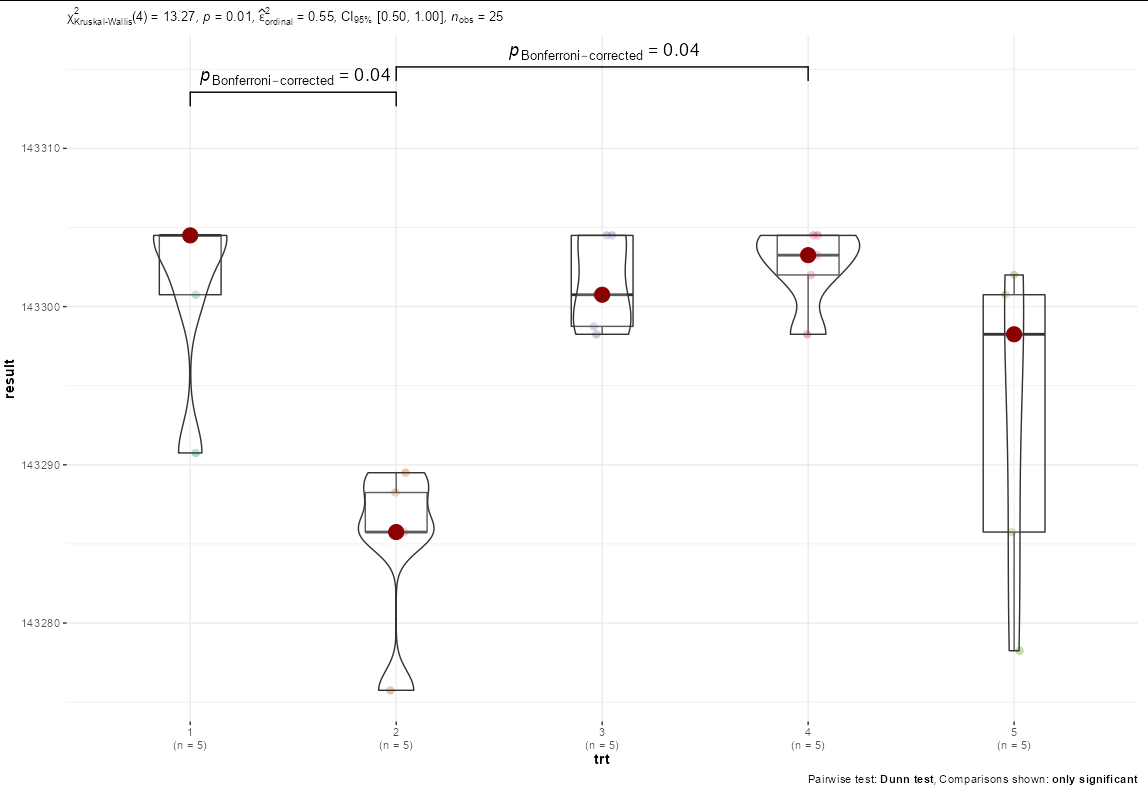
I am using a Kruskal-Wallis test to determine significant differences between treatments. Since this test is not an ANOVA, I cannot use the traditional post-hoc mean separation letters above the bar graph. There are some groups that are statistically different than others, and I would like to indicate the groups that are statistically different from one another on the graph.
Here is the data I am working with:
example <- data.frame(trt = c(5,
1,
2,
3,
4,
5,
2,
1,
3,
4,
1,
4,
5,
3,
2,
4,
1,
3,
5,
2,
4,
2,
1,
3,
5),
result=c(143278.25,
143290.75,
143275.75,
143298.25,
143298.25,
143285.75,
143285.75,
143304.5,
143304.5,
143302,
143300.75,
143303.25,
143298.25,
143304.5,
143285.75,
143304.5,
143304.5,
143300.75,
143302,
143288.25,
143304.5,
143289.5,
143304.5,
143298.75,
143300.75))
First, run the Kruskal-Wallace test:
library(FSA)
library(ggplot2)
library(dplyr)
kruskal.test(result ~ trt, data = example)
Results:
Kruskal-Wallis rank sum test
data: result by trt
Kruskal-Wallis chi-squared = 13.266, df = 4, p-value = 0.01005
These results indicate significant differences between groups, so the Dunn test will be used as a post-hoc test:
example$trt <- as.factor(example$trt)
dunnTest(result ~ trt, data = example, method = "bonferroni")
Results of this test indicate significance between groups 1-2, 2-3, and 2-4:
Comparison Z P.unadj P.adj
1 1 - 2 2.87420990 0.004050397 0.04050397
2 1 - 3 0.34838908 0.727548003 1.00000000
3 2 - 3 -2.52582083 0.011542833 0.11542833
4 1 - 4 -0.02177432 0.982627981 1.00000000
5 2 - 4 -2.89598422 0.003779714 0.03779714
6 3 - 4 -0.37016340 0.711260748 1.00000000
7 1 - 5 1.80726835 0.070720448 0.70720448
8 2 - 5 -1.06694156 0.285998228 1.00000000
9 3 - 5 1.45887927 0.144598340 1.00000000
10 4 - 5 1.82904267 0.067393217 0.67393217
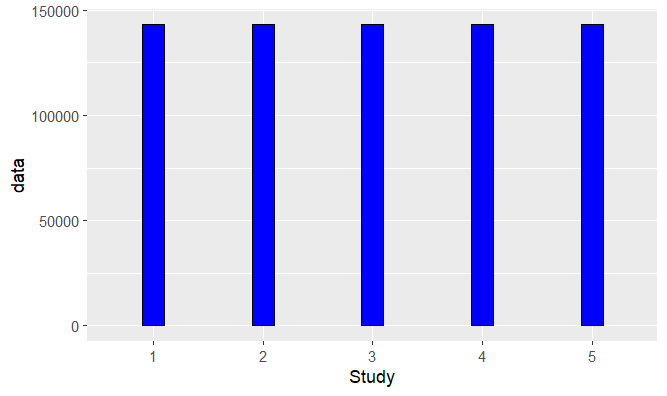
Generating the bar graph:
sem <- function(x) sd(x)/sqrt(length(x)) # Function for standard error of the mean
avg_data <- example %>%
group_by(trt) %>%
summarise(data = mean(result), .groups = 'drop')
sem_data <- example %>%
group_by(trt) %>%
summarise(data = sem(result), .groups = 'drop')
ggplot(avg_data, aes(x = trt, y = data))
geom_bar(stat="identity", width = 0.2, position = "dodge", col = "black", fill = "blue")
geom_errorbar(aes(ymin = data, ymax = data sem_data$data), width = 0.2, position = position_dodge(0.6))
xlab("Study") ylab("data")
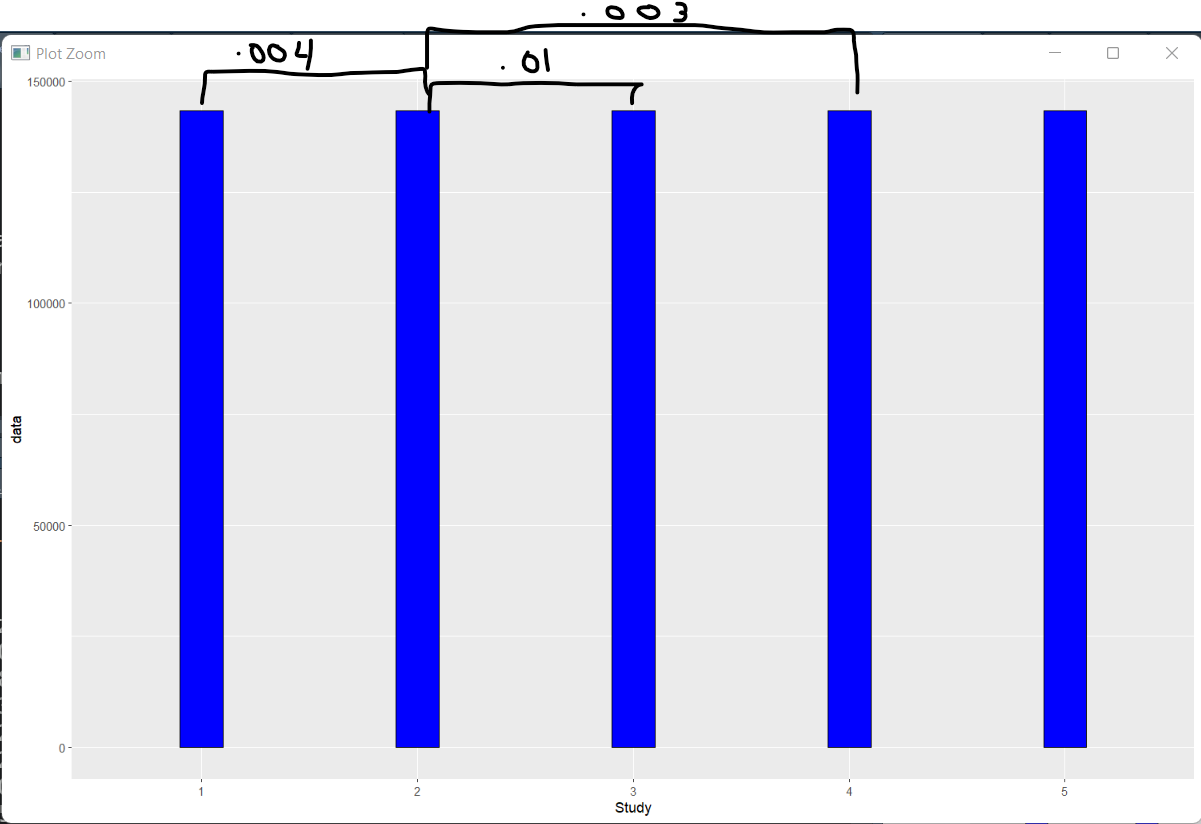
I would like to add the p-values for the significantly differnet groups with brackets above the bars, as illustrated in the picture below:
Is there a way that this can be done?
CodePudding user response:
I think a bar plot is a poor choice here. Because of the y axis scale, the bar heights all look the same. Showing only the mean of a variable using a bar plot also hides useful information about the distribution and sample size that could easily be displayed. This has led many people to advocate