I have a dataset composed of data with the same unit of measurement. Before making my pca, I centered my data using sklearn.preprocessing.StandardScaler(with_std=False).
I don't understand why but using the sklearn.decomposition.PCA.fit_transform(<my_dataframe>) method when I want to display a correlation circle I get two perfectly represented orthogonal variables, thus indicating that they are independent, but they are not. With a correlation matrix I observe perfectly that they are anti-correlated.
Through dint of research I came across the "
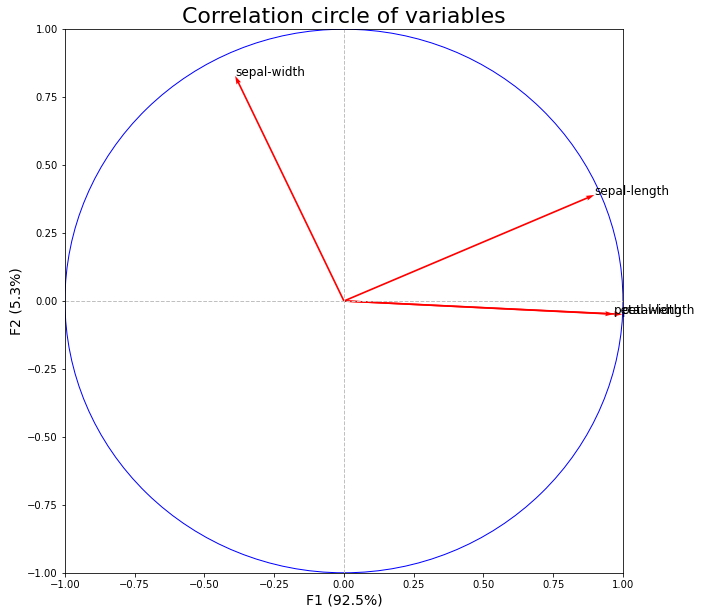
Here's the one I tinkered with to get my circle of correlations:
pcs = prince_pca.column_correlations(dataset)
pcs_0=pcs[0].to_numpy()
pcs_1=pcs[1].to_numpy()
pcs_coord = np.concatenate((pcs_0, pcs_1))
fig = plt.subplots(figsize=(10,10))
plt.xlim(-1,1)
plt.ylim(-1,1)
plt.quiver(np.zeros(pcs_0.shape[0]), np.zeros(pcs_1.shape[0]),
pcs_coord[:4], pcs_coord[4:], angles='xy', scale_units='xy', scale=1, color='r', width= 0.003)
for i, (x, y) in enumerate(zip(pcs_coord[:4], pcs_coord[4:])):
plt.text(x, y, pcs.index[i], fontsize=12)
circle = plt.Circle((0,0), 1, facecolor='none', edgecolor='b')
plt.gca().add_artist(circle)
plt.plot([-1,1],[0,0],color='silver',linestyle='--',linewidth=1)
plt.plot([0,0],[-1,1],color='silver',linestyle='--',linewidth=1)
plt.title("Correlation circle of variable", fontsize=22)
plt.xlabel('F{} ({}%)'.format(1, round(100*prince_pca.explained_inertia_[0],1)),
fontsize=14)
plt.ylabel('F{} ({}%)'.format(2, round(100*prince_pca.explained_inertia_[1],1)),
fontsize=14)
plt.show()
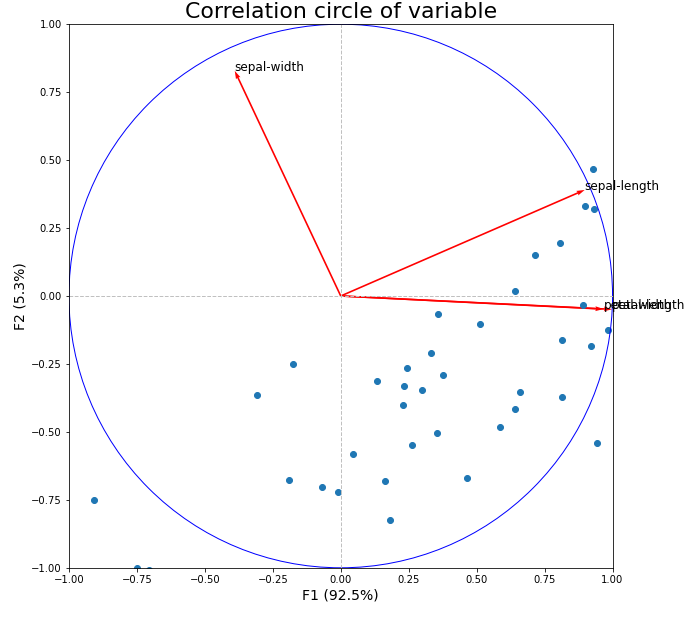
And finally here is the one that tries to bring together the circle of correlations as well as the main row coordinates graph from the "prince" package:
pcs = prince_pca.column_correlations(dataset)
pcs_0 = pcs[0].to_numpy()
pcs_1 = pcs[1].to_numpy()
pcs_coord = np.concatenate((pcs_0, pcs_1))
fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(111, aspect="equal")
plt.xlim(-1, 1)
plt.ylim(-1, 1)
plt.quiver(np.zeros(pcs_0.shape[0]),
np.zeros(pcs_1.shape[0]),
pcs_coord[:4],
pcs_coord[4:],
angles='xy',
scale_units='xy',
scale=1,
color='r',
width=0.003)
for i, (x, y) in enumerate(zip(pcs_coord[:4], pcs_coord[4:])):
plt.text(x, y, pcs.index[i], fontsize=12)
plt.scatter(
x=prince_pca.row_coordinates(dataset)[0],
y=prince_pca.row_coordinates(dataset)[1])
circle = plt.Circle((0, 0), 1, facecolor='none', edgecolor='b')
plt.gca().add_artist(circle)
plt.plot([-1, 1], [0, 0], color='silver', linestyle='--', linewidth=1)
plt.plot([0, 0], [-1, 1], color='silver', linestyle='--', linewidth=1)
plt.title("Correlation circle of variable", fontsize=22)
plt.xlabel('F{} ({}%)'.format(1,
round(100 * prince_pca.explained_inertia_[0],
1)),
fontsize=14)
plt.ylabel('F{} ({}%)'.format(2,
round(100 * prince_pca.explained_inertia_[1],
1)),
fontsize=14)
plt.show()
Bonus question: how to explain that the PCA class of sklearn does not calculate the correct coordinates for my variables when they are centered but not scaled? Any method to overcome this?
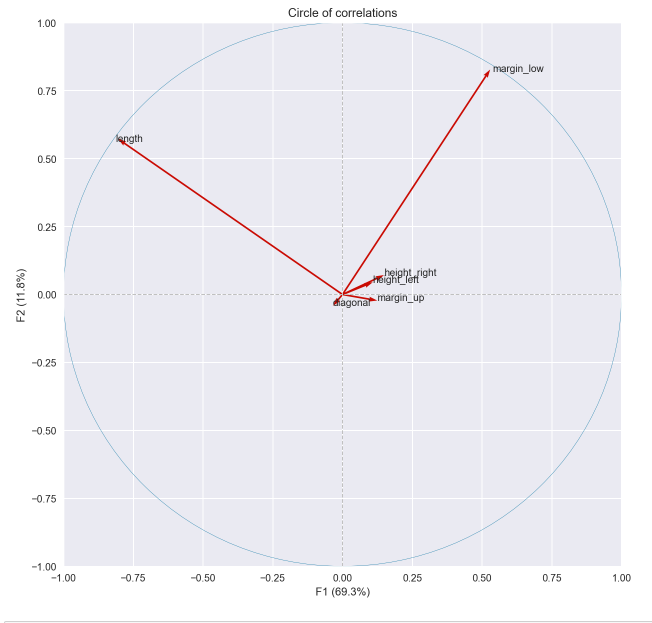
Here is the circle of correlations obtained by creating the pca object with sklearn where the "length" and "margin_low" variables appear as orthogonal:
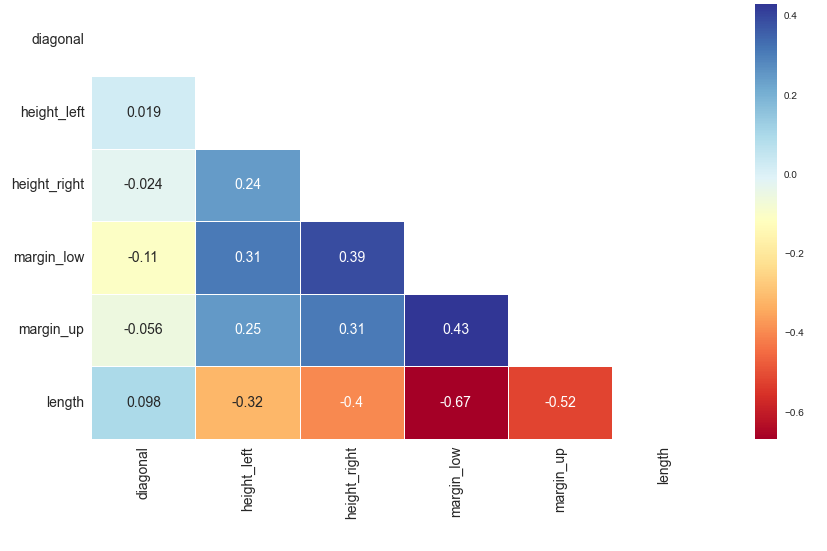
Here is the correlation matrix demonstrating the negative correlation between the "length" and "margin_low" variables:
CodePudding user response:
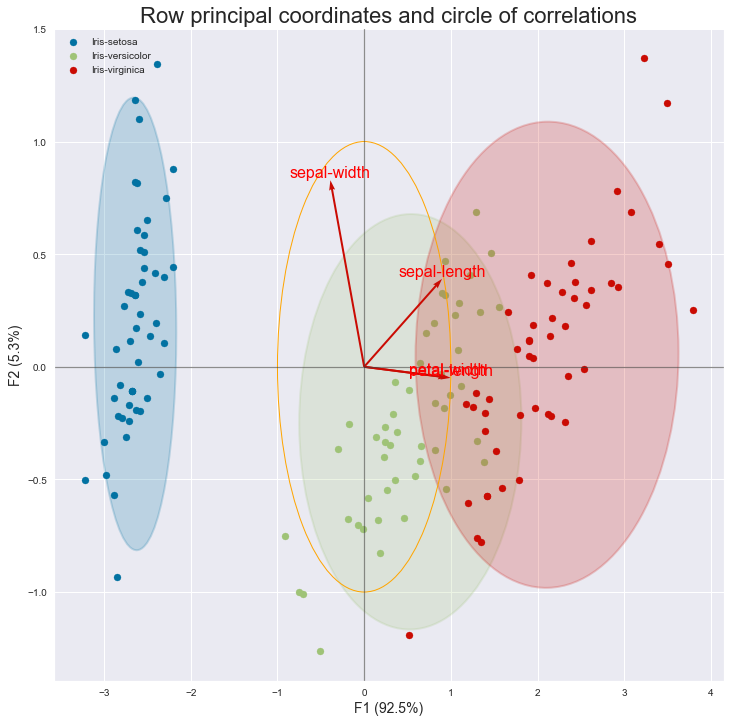
I managed to mix the two graphs.
Here is the code to display the graph combining the circle of correlations and the scatter with the rows:
import pandas as pd
import prince
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt
import numpy as np
# Import dataset
url = "https://archive.ics.uci.edu/ml/machine-learning-databases/iris/iris.data"
# Preparing the dataset
names = ['sepal-length', 'sepal-width', 'petal-length', 'petal-width', 'Class']
dataset = pd.read_csv(url, names=names)
dataset = dataset.set_index('Class')
# Preprocessing: centered but not scaled
sc = StandardScaler(with_std=False)
dataset = pd.DataFrame(sc.fit_transform(dataset),
index=dataset.index,
columns=dataset.columns)
# PCA setting
prince_pca = prince.PCA(n_components=2,
n_iter=3,
rescale_with_mean=True,
rescale_with_std=False,
copy=True,
check_input=True,
engine='auto',
random_state=42)
# PCA fiting
prince_pca = prince_pca.fit(dataset)
# Component coordinates
pcs = prince_pca.column_correlations(dataset)
# Row coordinates
pca_row_coord = prince_pca.row_coordinates(dataset).to_numpy()
# Preparing the colors for parameter 'c'
colors = dataset.T
# Display row coordinates
ax = prince_pca.plot_row_coordinates(dataset,
figsize=(12, 12),
x_component=0,
y_component=1,
labels=None,
color_labels=dataset.index,
ellipse_outline=True,
ellipse_fill=True,
show_points=True)
# We plot the vectors
plt.quiver(np.zeros(pcs.to_numpy().shape[0]),
np.zeros(pcs.to_numpy().shape[0]),
pcs[0],
pcs[1],
angles='xy',
scale_units='xy',
scale=1,
color='r',
width=0.003)
# Display the names of the variables
for i, (x, y) in enumerate(zip(pcs[0], pcs[1])):
if x >= xmin and x <= xmax and y >= ymin and y <= ymax:
plt.text(x,
y,
prince_pca.column_correlations(dataset).index[i],
fontsize=16,
ha="center",
va="bottom",
color="red")
# Display a circle
circle = plt.Circle((0, 0),
1,
facecolor='none',
edgecolor='orange',
linewidth=1)
plt.gca().add_artist(circle)
# Title
plt.title("Row principal coordinates and circle of correlations", fontsize=22)
# Display the percentage of inertia on each axis
plt.xlabel('F{} ({}%)'.format(1,
round(100 * prince_pca.explained_inertia_[0],
1)),
fontsize=14)
plt.ylabel('F{} ({}%)'.format(2,
round(100 * prince_pca.explained_inertia_[1],
1)),
fontsize=14)
# Display the grid to better read the values of the circle of correlations
plt.grid(visible=True)
plt.show()