I have the data as follows:
library(survival)
library(survminer)
data(aml)
aml$x <- as.character(aml$x)
aml[10,3] <- 'SuperMaintained'
aml[11,3] <- 'SuperMaintained'
aml[22,3] <- 'SuperMaintained'
aml[23,3] <- 'SuperMaintained'
aml$x <- factor(aml$x, levels = c('Nonmaintained','Maintained','SuperMaintained'))
fit <- survfit(Surv(time, status) ~ x, data = aml)
ggsurvplot(fit, conf.int = FALSE, surv.median.line = c('hv'), data = aml, pval = TRUE, risk.table = FALSE)
[![Survivalplot][1]][1]
res=pairwise_survdiff(Surv(time, status) ~ x, data = aml)
res$p.value
Output look like this
Nonmaintained Maintained
Maintained 0.085155296 NA
SuperMaintained 0.007303126 0.0851553
How do I add this table to the main survival plot?
CodePudding user response:
You can add a custom text annotate by adding a ggplot function to the "plot" (p1$plot) object saved in your ggsurvplot object. You can use the function annotate with geom=table thanks to the ggpmisc package and pass your table to the label argument like this:
library(survival)
library(survminer)
library(ggpmisc)
data("aml")
aml$x <- as.character(aml$x)
aml[10,3] <- 'SuperMaintained'
aml[11,3] <- 'SuperMaintained'
aml[22,3] <- 'SuperMaintained'
aml[23,3] <- 'SuperMaintained'
aml$x <- factor(aml$x, levels = c('Nonmaintained','Maintained','SuperMaintained'))
fit <- survfit(Surv(time, status) ~ x, data = aml)
res=pairwise_survdiff(Surv(time, status) ~ x, data = aml)
table <- res$p.value
p1 <- ggsurvplot(fit, conf.int = FALSE, surv.median.line = c('hv'), data = aml, pval = TRUE, risk.table = FALSE)
p1$plot
annotate(geom = "table", x = 140, y = 0.9, label = list(as.data.frame(table)))
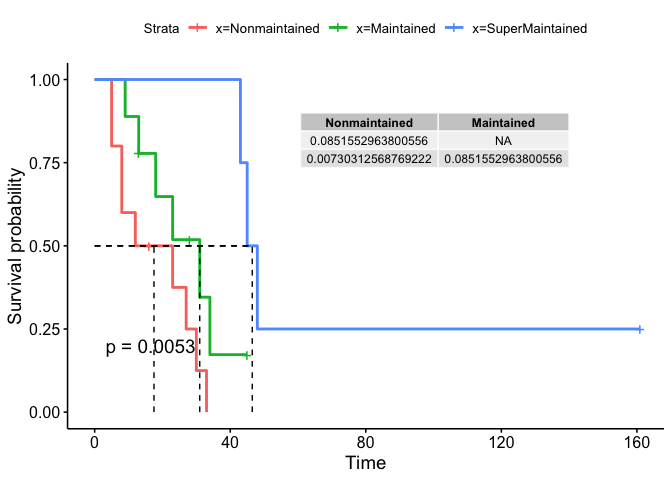
Created on 2022-08-19 with reprex v2.0.2
Please note: your "table" should be a tibble or data.frame, so I used as.data.frame().
Add row names to table
You can use rownames_to_column to include the rownames in your table like this:
# Create table for in plot
table <- as.data.frame(table)
table <- rownames_to_column(table)
colnames(table)[1] <- ""
p1 <- ggsurvplot(fit, conf.int = FALSE, surv.median.line = c('hv'), data = aml, pval = TRUE, risk.table = FALSE)
p1$plot
annotate(geom = "table", x = 140, y = 0.9, label = list(table))
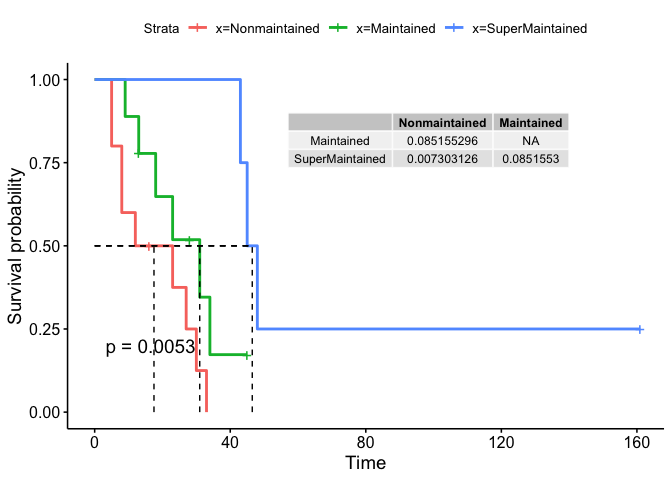
Created on 2022-08-19 with reprex v2.0.2
