I have been trying to find a solution to this, but have been unsuccessful as I am not 100% sure what is causing the issue. As this is the last little thing keeping me from finsihing this project, any ideas would be sooo much appreciated!
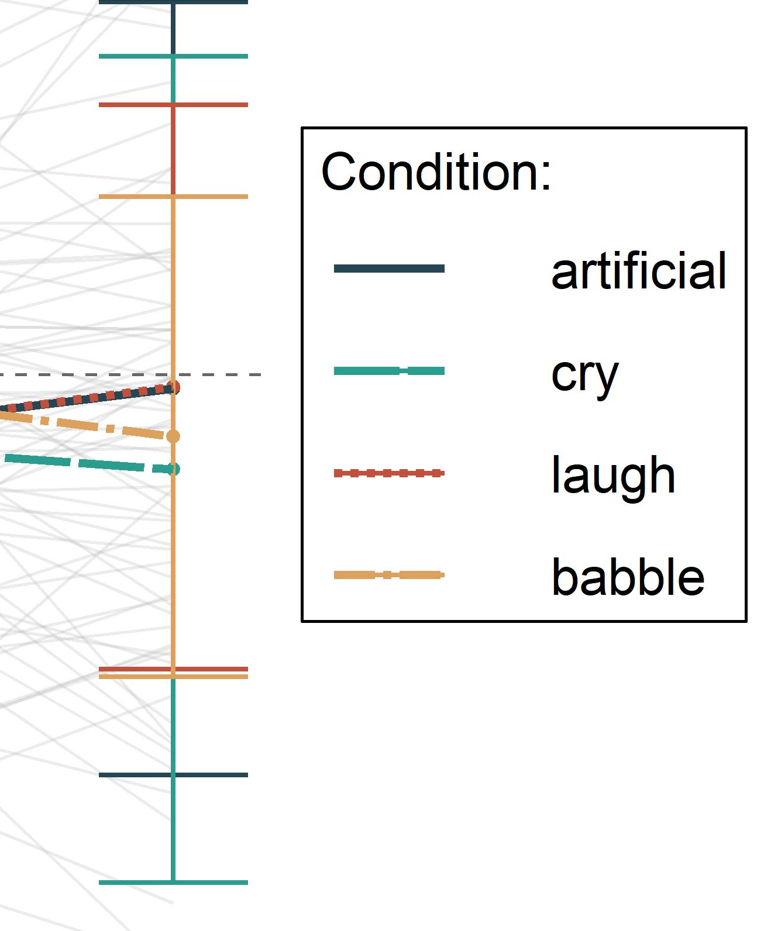
My ggplot legend looks like this:
As you can see, the linetype and colour are displayed combined, but there seems to be an additional solid line going through the linetypes, making them hard to read. I think this comes from an additional geom_line call to display the grey lines (raw data).
Here is my code minus the theme and labelling
ggplot(table_mean_a, mapping= aes(x = phase_bins, y = Mean))
geom_point(aes(x = phase_bins, y = Mean, colour = cond_f))
geom_line(df_plotting_a,
mapping = aes(x=phase_bins, y = mean_change_to_base, group=interaction(ID, cond_f)),
alpha = 0.3, size =0.5, colour = "grey", inherit.aes = FALSE)
geom_hline(yintercept=0, colour="#666766", linetype = "dashed")
geom_errorbar(subset(table_mean_a, phase_bins!= "baseline"),
mapping=aes(ymin=Mean-SD, ymax=Mean SD, colour = cond_f),
width=.4, size = 0.7)
geom_line(aes(group=as.numeric(cond_f), colour = cond_f, linetype=cond_f), size = 1.2)
facet_wrap(sample~., labeller=labeller(sample = labels), ncol=2, scales = "free_x")
scale_colour_manual(values=apa)
labs(colour = "Condition:", linetype = "Condition:")
guides(colour = guide_legend(override.aes = list(shape = NA, alpha = 1), byrow = TRUE))
scale_x_discrete(labels= c("base", "stim1", "stim2", "recovery", "break"), drop = T, expand = c(0,0.25))
scale_y_continuous(limits = c(-1,1), breaks = c(-1,-0.5,0,0.5,1),
labels = scales::number_format(accuracy = 1))
scale_linetype_manual(values=c("solid", "81", "11", "5111"))
Here is the data
table_mean_a <-
structure(list(cond_f = structure(c(1L, 1L, 1L, 1L, 1L, 2L, 2L,
2L, 2L, 2L), .Label = c("artificial", "cry", "laugh", "babble"
), class = "factor"), phase_bins = structure(c(2L, 3L, 4L, 5L,
6L, 2L, 3L, 4L, 5L, 6L), .Label = c("pre", "baseline", "stim_bin1",
"stim_bin2", "recovery", "break"), class = "factor"), age_group = c("adult",
"adult", "adult", "adult", "adult", "adult", "adult", "adult",
"adult", "adult"), sample = structure(c(3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L), .Label = c("UKi", "UGi", "UKa"), class = "factor"),
Mean = c(0, -0.0616666666666667, -0.143333333333333, -0.0983333333333328,
-0.0231666666666662, 0, -0.0949999999999997, -0.1625, -0.113333333333333,
-0.155833333333333), SD = c(0, 0.154096119467973, 0.24172866718288,
0.423777912706934, 0.633257128379362, 0, 0.13934894565432,
0.248908435688157, 0.350921926972951, 0.677239732274149),
Min. = c(0, -0.366666666666667, -0.583333333333337, -0.716666666666667,
-0.98, 0, -0.466666666666665, -0.650000000000003, -0.783333333333337,
-1.96), Max. = c(0, 0.283333333333334, 0.383333333333336,
1.08333333333334, 1.72666666666667, 0, 0.199999999999996,
0.233333333333333, 0.5, 0.846666666666667), count = c(20L,
20L, 20L, 20L, 20L, 20L, 20L, 20L, 20L, 20L)), row.names = c(NA,
-10L), groups = structure(list(cond_f = structure(c(1L, 1L, 1L,
1L, 1L, 2L, 2L, 2L, 2L, 2L), .Label = c("artificial", "cry",
"laugh", "babble"), class = "factor"), phase_bins = structure(c(2L,
3L, 4L, 5L, 6L, 2L, 3L, 4L, 5L, 6L), .Label = c("pre", "baseline",
"stim_bin1", "stim_bin2", "recovery", "break"), class = "factor"),
age_group = c("adult", "adult", "adult", "adult", "adult",
"adult", "adult", "adult", "adult", "adult"), .rows = structure(list(
1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L, 9L, 10L), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), row.names = c(NA, -10L), class = c("tbl_df",
"tbl", "data.frame"), .drop = TRUE), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"))
df_plotting_a <-
structure(list(ID = structure(c(69L, 69L, 69L, 69L, 69L), .Label = c("UG201",
"UG208", "UG209", "UG211", "UG215", "UG217", "UG219", "UG220",
"UG221", "UG222", "UG228", "UG243", "UG247", "UG254", "UG268",
"UG271", "UG272", "UG273", "UG274", "UG275", "UG280", "UG283",
"UG284", "UG286", "UG297", "UG299", "UG308", "UG310", "UG315",
"UG316", "UG330", "UG331", "UG334", "UG335", "UK103", "UK104",
"UK105", "UK106", "UK107", "UK108", "UK110", "UK111", "UK112",
"UK113", "UK115", "UK116", "UK117", "UK119", "UK122", "UK123",
"UK130", "UK132", "UK135", "UK136", "UK138", "UK139", "UK140",
"UK142", "UK145", "UK147", "UK150", "UK153", "UK155", "UK156",
"UK159", "UK160", "UK162", "UK164", "UKA102", "UKA103", "UKA104",
"UKA105", "UKA106", "UKA107", "UKA108", "UKA109", "UKA110", "UKA111",
"UKA112", "UKA113", "UKA114", "UKA115", "UKA116", "UKA117", "UKA119",
"UKA120", "UKA121", "UKA122"), class = "factor"), sex = structure(c(2L,
2L, 2L, 2L, 2L), .Label = c("f", "m"), class = "factor"), trial = structure(c(1L,
1L, 1L, 1L, 1L), .Label = c("1", "2", "3", "4"), class = "factor"),
cond_f = structure(c(4L, 4L, 4L, 4L, 4L), .Label = c("artificial",
"cry", "laugh", "babble"), class = "factor"), age_group = c("adult",
"adult", "adult", "adult", "adult"), stimulus = structure(c(7L,
7L, 7L, 7L, 7L), .Label = c("a1", "a2", "a3", "a4", "b1",
"b2", "b3", "b4", "c1", "c2", "c3", "c4", "l1", "l2", "l3",
"l4"), class = "factor"), phase_bins = structure(2:6, .Label = c("pre",
"baseline", "stim_bin1", "stim_bin2", "recovery", "break"
), class = "factor"), mean_change_to_base = c(0, -0.149999999999997,
-0.399999999999996, -0.299999999999996, -0.0733333333333306
), max_change_to_base = c(0, -0.333333333333329, -0.533333333333331,
-0.43333333333333, -0.333333333333329), sample = structure(c(3L,
3L, 3L, 3L, 3L), .Label = c("UKi", "UGi", "UKa"), class = "factor")), row.names = c(NA,
-5L), groups = structure(list(ID = structure(69L, .Label = c("UG201",
"UG208", "UG209", "UG211", "UG215", "UG217", "UG219", "UG220",
"UG221", "UG222", "UG228", "UG243", "UG247", "UG254", "UG268",
"UG271", "UG272", "UG273", "UG274", "UG275", "UG280", "UG283",
"UG284", "UG286", "UG297", "UG299", "UG308", "UG310", "UG315",
"UG316", "UG330", "UG331", "UG334", "UG335", "UK103", "UK104",
"UK105", "UK106", "UK107", "UK108", "UK110", "UK111", "UK112",
"UK113", "UK115", "UK116", "UK117", "UK119", "UK122", "UK123",
"UK130", "UK132", "UK135", "UK136", "UK138", "UK139", "UK140",
"UK142", "UK145", "UK147", "UK150", "UK153", "UK155", "UK156",
"UK159", "UK160", "UK162", "UK164", "UKA102", "UKA103", "UKA104",
"UKA105", "UKA106", "UKA107", "UKA108", "UKA109", "UKA110", "UKA111",
"UKA112", "UKA113", "UKA114", "UKA115", "UKA116", "UKA117", "UKA119",
"UKA120", "UKA121", "UKA122"), class = "factor"), trial = structure(1L, .Label = c("1",
"2", "3", "4"), class = "factor"), cond_f = structure(4L, .Label = c("artificial",
"cry", "laugh", "babble"), class = "factor"), .rows = structure(list(
1:5), ptype = integer(0), class = c("vctrs_list_of", "vctrs_vctr",
"list"))), row.names = c(NA, -1L), class = c("tbl_df", "tbl",
"data.frame"), .drop = TRUE), class = c("grouped_df", "tbl_df",
"tbl", "data.frame"))
CodePudding user response:
The legends from geom_errorbar and geom_line are overlapping together, therefore you see the solid line. You can use show.legend = FALSE in geom_errorbar to hide it.
Since some of your element used in ggplot is missing, here I only include the code for geom_errorbar.
geom_errorbar(subset(table_mean_a, phase_bins!= "baseline"),
mapping=aes(ymin=Mean-SD, ymax=Mean SD, colour = cond_f),
width=.4, size = 0.7, show.legend = F)