I can run ANOVA and TukeyHSD on my dataset, and the output can easily be saved to a txt format as such:
#AOV test
res.aov <- aov(variable_of_interest ~ Treatment*some_variable*some_parameter, data = dataframe)
#save output to text file
capture.output(TukeyHSD(res.aov), file="TUKEY_outfile.txt")
However, the output is pretty ugly in my opinion, and it takes a lot of time to get it into Excel and format it into a pretty format. Is there a way to directly print the outputs to a nice table format in for example excel or .pdf? I saw a LATEX solution, but it was unfortunately very complicated.
As a working example, the following code could be used:
fm1 <- aov(breaks ~ wool tension, data = warpbreaks)
capture.output(TukeyHSD(fm1), file="example_tukey.txt")
CodePudding user response:
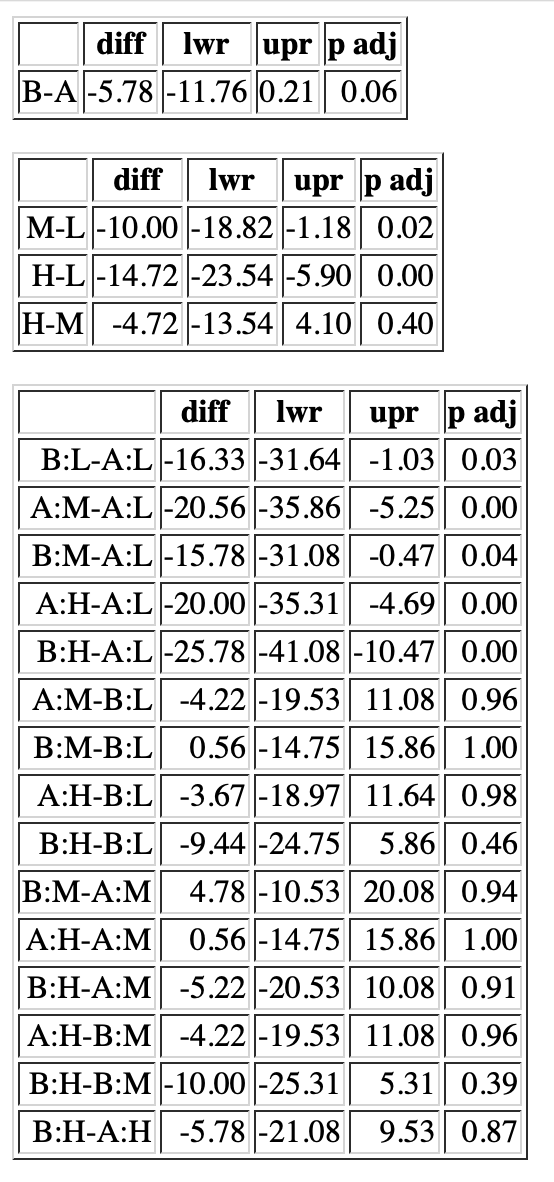
Here is a possible solution. If you plan to move code/output/tables/graphics from R to pdf/html/latex format files regularly, you should invest in the time to learn Rmarkdown or Bookdown which are specifically designed exactly for this purpose. If you just want to export a few tables to html or latex, the package xtable may work for you. The following code uses the example on the TukeyHSD manual page to illustrate producing an html file containing three tables:
fm1 <- aov(breaks ~ wool * tension, data = warpbreaks)
TK <- TukeyHSD(fm1)
print(xtable(TK$wool), type="html", file="TukeyTables.html")
cat("<p></p>", file="TukeyTables.html", append=TRUE)
print(xtable(TK$tension), type="html", file="TukeyTables.html", append=TRUE)
cat("<p></p>", file="TukeyTables.html", append=TRUE)
print(xtable(TK$'wool:tension'), type="html", file="TukeyTables.html", append=TRUE)
This code produces an html file with three tables will look something like the figure. There are many arguments in xtable function that give you substantial control over the formatting, but the package is really designed for latex.