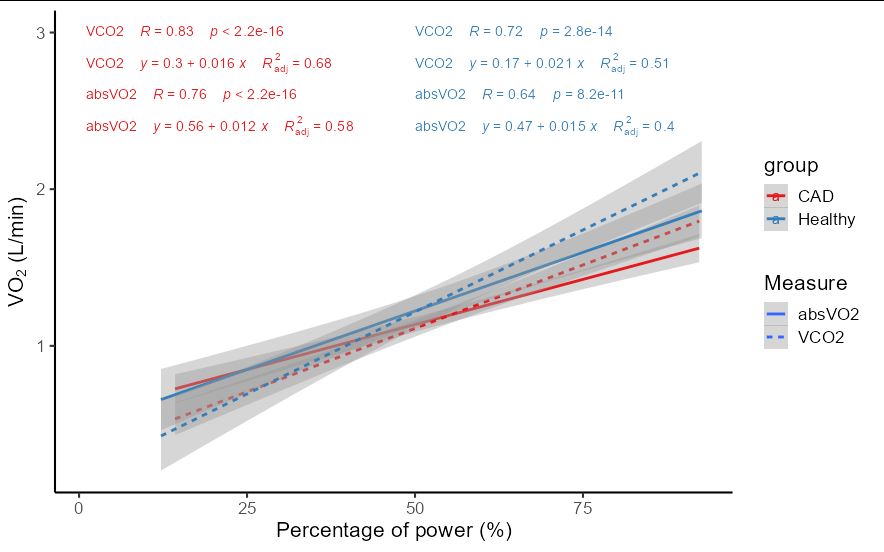
I've got 2 plots showing VO2 and VCO2 according percent_power.
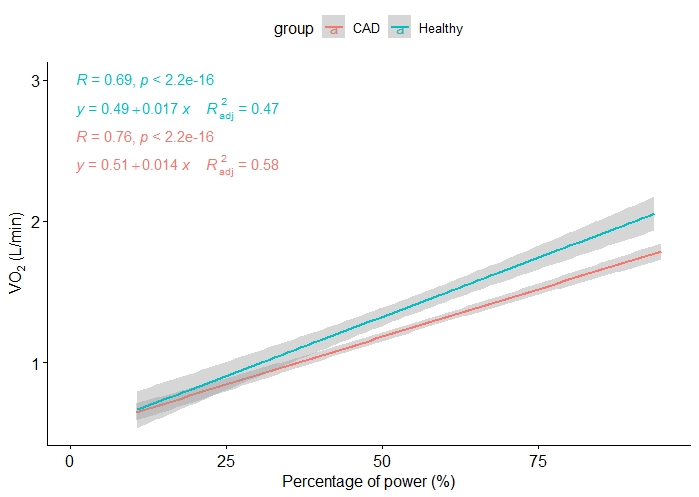
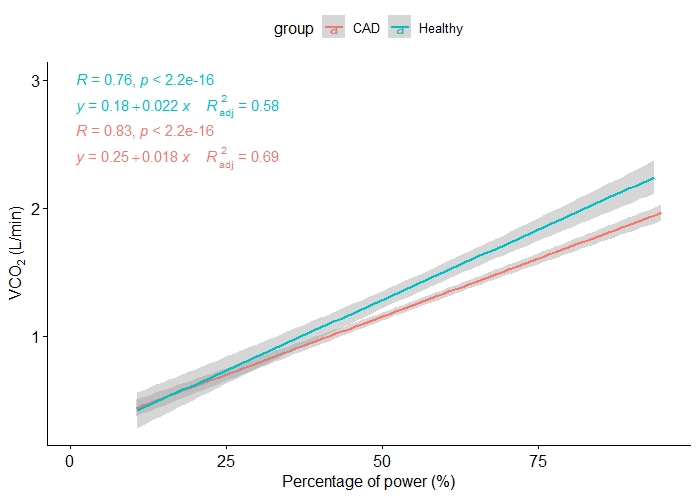
I would like to create 2 plots:
- One where
VO2andVCO2are combined since theirYaxes have a similar scale, but only inhealthygroup - Same, but in
CADgroup only.
They can be distinguished by the type of line or color. What I have tried, is to combine them but I still have 2 different plots.
Here are the code for the 2 plots:
##VO2
df %>%
filter(percent_power < 100 & temps == "1") %>%
ggscatter(x = "percent_power", y = "absVO2", color = "group", point = F)
stat_cor(aes(color = group), label.x = 1, label.y = c(2.6,3))
stat_regline_equation(label.x = 1, label.y = c(2.4,2.8),
formula = y ~ x,
aes(color = group, label = paste(..eq.label.., ..adj.rr.label.., sep = "~~~~")),)
geom_smooth(aes(colour=group), method = "lm", formula = y ~ x)
xlab("Percentage of power (%)")
ylab(expression(paste("V", O[2]," (L/min)")))
##VCO2
df %>%
filter(percent_power < 100 & temps == "1") %>%
ggscatter(x = "percent_power", y = "VCO2", color = "group", point = F)
stat_cor(aes(color = group), label.x = 1, label.y = c(2.6,3))
stat_regline_equation(label.x = 1, label.y = c(2.4,2.8),
formula = y ~ x,
aes(color = group, label = paste(..eq.label.., ..adj.rr.label.., sep = "~~~~")),)
geom_smooth(aes(colour=group), method = "lm", formula = y ~ x)
xlab("Percentage of power (%)")
ylab(expression(paste("VC", O[2]," (L/min)")))
Thank you very much!
And here are my data to work on:
structure(list(id = c("AC12-PRD-C1", "AC12-PRD-C1", "AC12-PRD-C1",
"AC12-PRD-C1", "AC12-PRD-C1", "AC12-PRD-C1", "AC12-PRD-C1", "AL13-PRD-C1",
"AL13-PRD-C1", "AL13-PRD-C1", "AL13-PRD-C1", "AL13-PRD-C1", "AL13-PRD-C1",
"AL13-PRD-C1", "AL13-PRD-C1", "BM06-PRD-S1", "BM06-PRD-S1", "BM06-PRD-S1",
"BM06-PRD-S1", "BM06-PRD-S1", "BM06-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1",
"CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1",
"CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1", "CB19-PRD-S1",
"CC14-PRD-S1", "CC14-PRD-S1", "CC14-PRD-S1", "CC14-PRD-S1", "CC14-PRD-S1",
"CC14-PRD-S1", "CC14-PRD-S1", "CC14-PRD-S1", "DA03-PRD-C1", "DA03-PRD-C1",
"DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1",
"DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1", "DA03-PRD-C1",
"DA24-PRD-S1", "DA24-PRD-S1", "DA24-PRD-S1", "DA24-PRD-S1", "DA24-PRD-S1",
"DA24-PRD-S1", "DA24-PRD-S1", "DB22-PRD-S1", "DB22-PRD-S1", "DB22-PRD-S1",
"DB22-PRD-S1", "DB22-PRD-S1", "DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1",
"DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1",
"DB42-PRD-C1", "DB42-PRD-C1", "DB42-PRD-C1", "DL18-PRD-S1", "DL18-PRD-S1",
"DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1",
"DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1", "DL18-PRD-S1",
"DL18-PRD-S1", "DR15-PRD-C1", "DR15-PRD-C1", "DR15-PRD-C1", "DR15-PRD-C1",
"DR15-PRD-C1", "DR15-PRD-C1", "DR15-PRD-C1", "DR15-PRD-C1", "DT08-PRD-S1",
"DT08-PRD-S1", "DT08-PRD-S1", "DT08-PRD-S1", "DT08-PRD-S1", "DT08-PRD-S1",
"DT08-PRD-S1", "DT08-PRD-S1", "FB44-PRD-C1", "FB44-PRD-C1", "FB44-PRD-C1",
"FB44-PRD-C1", "FB44-PRD-C1", "FB44-PRD-C1", "FB44-PRD-C1", "FG33-PRD-C1",
"FG33-PRD-C1", "FG33-PRD-C1", "FG33-PRD-C1", "FG33-PRD-C1", "FG33-PRD-C1",
"FG33-PRD-C1", "FG33-PRD-C1", "FG33-PRD-C1", "FG33-PRD-C1", "FG33-PRD-C1",
"FG33-PRD-C1", "FG40-PRD-C1", "FG40-PRD-C1", "FG40-PRD-C1", "FG40-PRD-C1",
"FG40-PRD-C1", "GC03-PRD-S1", "GC03-PRD-S1", "GC03-PRD-S1", "GC03-PRD-S1",
"GC03-PRD-S1", "GC03-PRD-S1", "GC03-PRD-S1", "GC03-PRD-S1", "GG30-PRD-C1",
"GG30-PRD-C1", "GG30-PRD-C1", "GG30-PRD-C1", "GG30-PRD-C1", "GG30-PRD-C1",
"GG30-PRD-C1", "GG30-PRD-C1", "GH05-PRD-C1", "GH05-PRD-C1", "GH05-PRD-C1",
"GH05-PRD-C1", "GH05-PRD-C1", "GH05-PRD-C1", "GH05-PRD-C1", "GH05-PRD-C1",
"GH05-PRD-C1", "GH05-PRD-C1", "GL05-PRD-S1", "GL05-PRD-S1", "GL05-PRD-S1",
"GL05-PRD-S1", "GL05-PRD-S1", "GL05-PRD-S1", "GL05-PRD-S1", "GL05-PRD-S1",
"GL05-PRD-S1", "GL05-PRD-S1", "GL05-PRD-S1", "GL05-PRD-S1", "GP22-PRD-C1",
"GP22-PRD-C1", "GP22-PRD-C1", "GP22-PRD-C1", "GP22-PRD-C1", "GP22-PRD-C1",
"GP22-PRD-C1", "GP22-PRD-C1", "GP22-PRD-C1", "GP28-PRD-C1", "GP28-PRD-C1",
"GP28-PRD-C1", "GP28-PRD-C1", "GP28-PRD-C1", "GP28-PRD-C1", "GP28-PRD-C1",
"GP28-PRD-C1", "GP28-PRD-C1", "GP28-PRD-C1", "GP28-PRD-C1", "GT04-PRD-S1",
"GT04-PRD-S1", "GT04-PRD-S1", "GT04-PRD-S1", "GT04-PRD-S1", "GT04-PRD-S1",
"GT04-PRD-S1", "GT21-PRD-S1", "GT21-PRD-S1", "GT21-PRD-S1", "GT21-PRD-S1",
"GT21-PRD-S1", "GT21-PRD-S1", "GT21-PRD-S1", "GT21-PRD-S1"),
power = c(25L, 40L, 55L, 70L, 85L, 100L, 115L, 25L, 40L,
55L, 70L, 85L, 100L, 115L, 130L, 25L, 40L, 55L, 70L, 85L,
100L, 25L, 40L, 55L, 70L, 85L, 100L, 115L, 130L, 145L, 160L,
175L, 190L, 20L, 30L, 40L, 50L, 60L, 70L, 80L, 90L, 20L,
30L, 40L, 50L, 60L, 70L, 80L, 90L, 100L, 110L, 120L, 130L,
25L, 40L, 55L, 70L, 85L, 100L, 115L, 25L, 40L, 55L, 70L,
85L, 25L, 40L, 55L, 70L, 85L, 100L, 115L, 130L, 145L, 160L,
175L, 25L, 40L, 55L, 70L, 85L, 100L, 115L, 130L, 145L, 160L,
175L, 190L, 205L, 20L, 30L, 40L, 50L, 60L, 70L, 80L, 90L,
50L, 60L, 70L, 80L, 90L, 100L, 110L, 120L, 20L, 30L, 40L,
50L, 60L, 70L, 80L, 20L, 30L, 40L, 50L, 60L, 70L, 80L, 90L,
100L, 110L, 120L, 130L, 20L, 30L, 40L, 50L, 60L, 25L, 40L,
55L, 70L, 85L, 100L, 115L, 130L, 25L, 40L, 55L, 70L, 85L,
100L, 115L, 130L, 20L, 30L, 40L, 50L, 60L, 70L, 80L, 90L,
100L, 110L, 40L, 60L, 80L, 100L, 120L, 140L, 160L, 180L,
200L, 220L, 240L, 260L, 25L, 40L, 55L, 70L, 85L, 100L, 115L,
130L, 145L, 25L, 40L, 55L, 70L, 85L, 100L, 115L, 130L, 145L,
160L, 175L, 60L, 80L, 100L, 120L, 140L, 160L, 180L, 25L,
40L, 55L, 70L, 85L, 100L, 115L, 130L), absVO2 = c(0.739,
0.81975, 0.95125, 1.07525, 1.199, 1.34575, 1.49775, 0.66125,
0.7485, 0.855, 0.9845, 1.1495, 1.3465, 1.451, 1.5985, 0.61675,
0.717, 0.86275, 0.98575, 1.13, 1.262, 0.8835, 0.94575, 1.08125,
1.244, 1.30475, 1.4735, 1.679, 1.79075, 1.96, 2.0405, 2.34425,
2.4435, 0.5925, 0.661, 0.7435, 0.87875, 0.9435, 0.99675,
1.11425, 1.20275, 0.9255, 1.02925, 1.11675, 1.19725, 1.24775,
1.42625, 1.54225, 1.59425, 1.69675, 1.776, 1.94525, 2.0395,
0.535, 0.5845, 0.76875, 0.982, 1.09975, 1.259, 1.3265, 0.7205,
0.86825, 0.9235, 1.01075, 1.17275, 0.7435, 0.76575, 0.96075,
1.0975, 1.21975, 1.34525, 1.50625, 1.6755, 1.86325, 2.0465,
2.1395, 0.613, 0.85525, 0.98, 1.076, 1.287, 1.4615, 1.59325,
1.6965, 1.884, 1.998, 2.1425, 2.31275, 2.474, 0.81075, 0.8035,
0.8645, 1.076, 1.119, 1.255, 1.33825, 1.499, 0.8105, 0.8865,
1.03725, 1.134, 1.26675, 1.36275, 1.4455, 1.52875, 0.64675,
0.6645, 0.742, 0.81675, 0.983, 1.06875, 1.155, 1.02475, 1.0335,
1.09525, 1.21675, 1.283, 1.39025, 1.438, 1.575, 1.63075,
1.8005, 1.95575, 2.047, 0.708, 0.7245, 0.82675, 0.924, 0.99275,
0.6775, 0.69625, 0.806, 0.95825, 1.07425, 1.2515, 1.37325,
1.62675, 0.79325, 0.841, 0.97275, 1.04925, 1.16925, 1.41225,
1.53125, 1.65475, 0.7105, 0.74875, 0.85225, 0.92375, 1.05575,
1.155, 1.228, 1.33625, 1.4605, 1.5715, 1.02625, 1.23, 1.326,
1.57525, 1.7225, 1.907, 2.13825, 2.35175, 2.52325, 2.75525,
2.95425, 3.117, 0.727, 0.76075, 0.9335, 1.0545, 1.229, 1.37075,
1.44675, 1.59225, 1.7365, 0.99625, 0.991, 1.199, 1.20625,
1.4295, 1.52425, 1.587, 1.71075, 1.88425, 1.9755, 2.10025,
1.279, 1.41725, 1.547, 1.8155, 1.976, 2.1635, 2.3375, 0.80825,
0.87425, 1.01375, 1.1475, 1.286, 1.432, 1.56, 1.748), VCO2 = c(0.667,
0.715, 0.854, 1.047, 1.25125, 1.5185, 1.76025, 0.5805, 0.66425,
0.79425, 0.99925, 1.23475, 1.48375, 1.65775, 1.89075, 0.58225,
0.6595, 0.8275, 1.0455, 1.30525, 1.6115, 0.80475, 0.829,
0.958, 1.14275, 1.19925, 1.42225, 1.68475, 1.8795, 2.13525,
2.2835, 2.7605, 2.9685, 0.54825, 0.60325, 0.66025, 0.77325,
0.89375, 0.98525, 1.16275, 1.327, 0.781, 0.8775, 0.972, 1.05425,
1.1175, 1.353, 1.5075, 1.61425, 1.76925, 1.8675, 2.17975,
2.3575, 0.45075, 0.47775, 0.6545, 0.95525, 1.18325, 1.497,
1.725, 0.58275, 0.746, 0.7955, 0.91, 1.13175, 0.6205, 0.60475,
0.791, 0.963, 1.10725, 1.30025, 1.56775, 1.804, 2.1645, 2.434,
2.67125, 0.542, 0.76325, 0.89, 1.081, 1.2545, 1.4735, 1.68575,
1.83225, 2.1025, 2.31325, 2.52175, 2.755, 3.117, 0.72925,
0.7275, 0.77375, 1.02925, 1.1305, 1.33425, 1.48125, 1.77475,
0.70875, 0.839, 0.9905, 1.206, 1.4965, 1.649, 1.7875, 1.9635,
0.6755, 0.67525, 0.751, 0.86925, 1.06025, 1.23525, 1.4025,
0.871, 0.888, 0.97825, 1.11275, 1.23175, 1.3465, 1.4595,
1.60625, 1.74075, 2.029, 2.37675, 2.64675, 0.6175, 0.64475,
0.76425, 0.9405, 1.12525, 0.5545, 0.566, 0.69775, 0.8575,
1.014, 1.2, 1.42375, 1.878, 0.61675, 0.6825, 0.82925, 0.98575,
1.14725, 1.45, 1.6995, 1.92425, 0.6365, 0.67, 0.7485, 0.81925,
0.95025, 1.08175, 1.1795, 1.3525, 1.53, 1.67925, 0.73375,
0.90225, 1.0115, 1.3055, 1.498, 1.78, 2.0795, 2.436, 2.701,
3.06675, 3.38925, 3.68425, 0.6335, 0.681, 0.8515, 1.0125,
1.2365, 1.44175, 1.613, 1.8165, 1.99725, 0.82575, 0.83625,
1.03175, 1.06925, 1.3575, 1.549, 1.7125, 1.91125, 2.06925,
2.17725, 2.33825, 1.3015, 1.4475, 1.61525, 2.0795, 2.39475,
2.6085, 2.9635, 0.73725, 0.812, 0.9935, 1.1605, 1.361, 1.60875,
1.78375, 2.08475), percent_power = c(21.7391304347826, 34.7826086956522,
47.8260869565217, 60.8695652173913, 73.9130434782609, 86.9565217391304,
100, 19.2307692307692, 30.7692307692308, 42.3076923076923,
53.8461538461538, 65.3846153846154, 76.9230769230769, 88.4615384615385,
100, 25, 40, 55, 70, 85, 100, 13.1578947368421, 21.0526315789474,
28.9473684210526, 36.8421052631579, 44.7368421052632, 52.6315789473684,
60.5263157894737, 68.4210526315789, 76.3157894736842, 84.2105263157895,
92.1052631578947, 100, 22.2222222222222, 33.3333333333333,
44.4444444444444, 55.5555555555556, 66.6666666666667, 77.7777777777778,
88.8888888888889, 100, 15.3846153846154, 23.0769230769231,
30.7692307692308, 38.4615384615385, 46.1538461538462, 53.8461538461538,
61.5384615384615, 69.2307692307692, 76.9230769230769, 84.6153846153846,
92.3076923076923, 100, 21.7391304347826, 34.7826086956522,
47.8260869565217, 60.8695652173913, 73.9130434782609, 86.9565217391304,
100, 29.4117647058824, 47.0588235294118, 64.7058823529412,
82.3529411764706, 100, 14.2857142857143, 22.8571428571429,
31.4285714285714, 40, 48.5714285714286, 57.1428571428571,
65.7142857142857, 74.2857142857143, 82.8571428571429, 91.4285714285714,
100, 12.1951219512195, 19.5121951219512, 26.8292682926829,
34.1463414634146, 41.4634146341463, 48.780487804878, 56.0975609756098,
63.4146341463415, 70.7317073170732, 78.0487804878049, 85.3658536585366,
92.6829268292683, 100, 22.2222222222222, 33.3333333333333,
44.4444444444444, 55.5555555555556, 66.6666666666667, 77.7777777777778,
88.8888888888889, 100, 41.6666666666667, 50, 58.3333333333333,
66.6666666666667, 75, 83.3333333333333, 91.6666666666667,
100, 25, 37.5, 50, 62.5, 75, 87.5, 100, 15.3846153846154,
23.0769230769231, 30.7692307692308, 38.4615384615385, 46.1538461538462,
53.8461538461538, 61.5384615384615, 69.2307692307692, 76.9230769230769,
84.6153846153846, 92.3076923076923, 100, 33.3333333333333,
50, 66.6666666666667, 83.3333333333333, 100, 19.2307692307692,
30.7692307692308, 42.3076923076923, 53.8461538461538, 65.3846153846154,
76.9230769230769, 88.4615384615385, 100, 19.2307692307692,
30.7692307692308, 42.3076923076923, 53.8461538461538, 65.3846153846154,
76.9230769230769, 88.4615384615385, 100, 18.1818181818182,
27.2727272727273, 36.3636363636364, 45.4545454545455, 54.5454545454545,
63.6363636363636, 72.7272727272727, 81.8181818181818, 90.9090909090909,
100, 15.3846153846154, 23.0769230769231, 30.7692307692308,
38.4615384615385, 46.1538461538462, 53.8461538461538, 61.5384615384615,
69.2307692307692, 76.9230769230769, 84.6153846153846, 92.3076923076923,
100, 17.2413793103448, 27.5862068965517, 37.9310344827586,
48.2758620689655, 58.6206896551724, 68.9655172413793, 79.3103448275862,
89.6551724137931, 100, 14.2857142857143, 22.8571428571429,
31.4285714285714, 40, 48.5714285714286, 57.1428571428571,
65.7142857142857, 74.2857142857143, 82.8571428571429, 91.4285714285714,
100, 33.3333333333333, 44.4444444444444, 55.5555555555556,
66.6666666666667, 77.7777777777778, 88.8888888888889, 100,
19.2307692307692, 30.7692307692308, 42.3076923076923, 53.8461538461538,
65.3846153846154, 76.9230769230769, 88.4615384615385, 100
), group = c("CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD", "CAD",
"CAD", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy", "Healthy", "Healthy",
"Healthy", "Healthy", "Healthy", "Healthy")), class = c("grouped_df",
"tbl_df", "tbl", "data.frame"), row.names = c(NA, -202L), groups = structure(list(
id = c("AC12-PRD-C1", "AL13-PRD-C1", "BM06-PRD-S1", "CB19-PRD-S1",
"CC14-PRD-S1", "DA03-PRD-C1", "DA24-PRD-S1", "DB22-PRD-S1",
"DB42-PRD-C1", "DL18-PRD-S1", "DR15-PRD-C1", "DT08-PRD-S1",
"FB44-PRD-C1", "FG33-PRD-C1", "FG40-PRD-C1", "GC03-PRD-S1",
"GG30-PRD-C1", "GH05-PRD-C1", "GL05-PRD-S1", "GP22-PRD-C1",
"GP28-PRD-C1", "GT04-PRD-S1", "GT21-PRD-S1"), .rows = structure(list(
1:7, 8:15, 16:21, 22:33, 34:41, 42:53, 54:60, 61:65,
66:76, 77:89, 90:97, 98:105, 106:112, 113:124, 125:129,
130:137, 138:145, 146:155, 156:167, 168:176, 177:187,
188:194, 195:202), ptype = integer(0), class = c("vctrs_list_of",
"vctrs_vctr", "list"))), class = c("tbl_df", "tbl", "data.frame"
), row.names = c(NA, -23L), .drop = TRUE))
CodePudding user response:
You really don't need two ggscatter plots here. You could do this all as a single ggplot with less code:
df %>%
filter(percent_power < 100) %>%
tidyr::pivot_longer(absVO2:VCO2, names_to = 'Measure') %>%
mutate(groups = interaction(Measure, group, sep = ':')) %>%
ggplot(aes(percent_power, value, color = group, linetype = Measure))
geom_smooth(aes(group = groups), method = 'lm', formula = y~x)
stat_cor(aes(group = groups,
label = paste(..linetype.., ..r.label.., ..p.label..,
sep = "~~~~"),
color = group),
label.x = c(1, 1, 50, 50), label.y = c(2.6,3, 2.6, 3))
stat_regline_equation(label.x = c(1, 1, 50, 50),
label.y = c(2.4, 2.8, 2.4, 2.8),
formula = y ~ x,
aes(color = group,
label = paste(..linetype.., ..eq.label..,
..adj.rr.label..,
sep = "~~~~")))
xlab("Percentage of power (%)")
ylab(expression(paste("V", O[2]," (L/min)")))
scale_color_brewer(palette = 'Set1')
theme_classic(base_size = 16)