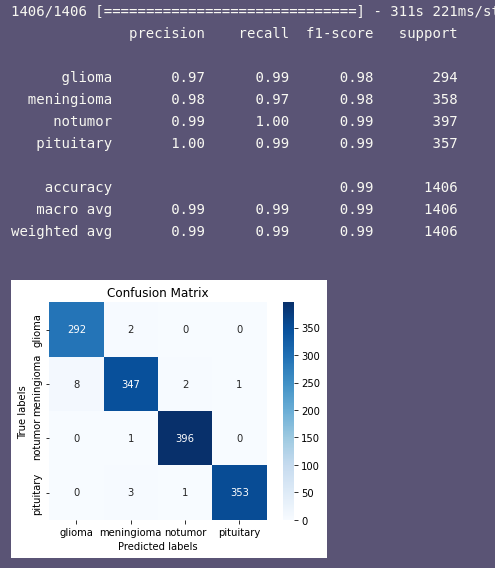
I am working on a tumor classification problem where I have 4 classes [glioma(1624 sample), meningioma(1645 sample), notumor(2000 sample) and pituitary(1757 sample)] . I am having problems with my saved model, the confusion matrix is showing nearly perfect results no bias or anything, when I do
model.evaluate(test_set)
I get : categorical_accuracy: 0.9872 [0.06456023454666138, 0.9871976971626282]
But when I try to do predictions all the results are wrong ( even when I purposly inject training data for the model to predict) I wrote this function for the prediction :
def predict_on_one_image(model,image_path):
img = imread(image_path)
img = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)
img = np.expand_dims(np.stack((img,)*3, axis=-1), axis=0).astype(np.float32)
p = model6.predict(img)
return {'class':p.argmax(axis=-1)[0],'probability':p}
my labels are :
{'glioma': 0, 'meningioma': 1, 'notumor': 2, 'pituitary': 3}
and if I try to predict the class of an image from the training set [ original class = glioma]
predict_on_one_image(model6,'path_here/training/glioma/Te-gl_0016.jpg' )
I get the class prediction as notumor
{'class': 2, 'probability': array([[0., 0., 1., 0.]], dtype=float32)}
I am doubting that its biasing to notumor because it has the most data ( class imbalance problem), but is there anyway I can get right predictions without having to run the model again?
CodePudding user response:
Basically, I needed to Process the images for prediction in the same way that I processed them for training ( I rescaled them using ImageDataGenerator)
I added these few lines of code, and it's predicting properly :
img = imread('path_here/testing/notumor/Te-no_0164.jpg')
x = asarray(img)
x = x / 255.
x = np.expand_dims(x, axis=0)
images = np.vstack([x])
classes = model6.predict(images, batch_size=10)
print('Predicted class is :')
print(np.argmax(classes))
Credits to: This comment