This is my data frame a subset of my big one as an example
dput(eee)
structure(list(interactome = c("HINT-binary", "HINT-binary",
"HINT-binary", "HINT-binary", "HINT-binary", "HINT-binary", "HINT-comp",
"HINT-comp", "HINT-comp", "HINT-comp", "HINT-comp", "HINT-comp",
"InBioMap", "InBioMap", "InBioMap", "InBioMap", "InBioMap", "InBioMap",
"Menche-2015", "Menche-2015", "Menche-2015", "Menche-2015", "Menche-2015",
"Menche-2015"), class = c("observed", "rewired", "rewired", "rewired",
"rewired", "rewired", "observed", "rewired", "rewired", "rewired",
"rewired", "rewired", "observed", "rewired", "rewired", "rewired",
"rewired", "rewired", "observed", "rewired", "rewired", "rewired",
"rewired", "rewired"), PPI = c(844L, 609L, 591L, 593L, 590L,
608L, 1329L, 874L, 872L, 864L, 807L, 855L, 7077L, 5049L, 5051L,
5025L, 4975L, 5014L, 2445L, 1673L, 1652L, 1716L, 1712L, 1683L
), LCC = c(290L, 191L, 188L, 214L, 183L, 215L, 401L, 346L, 365L,
366L, 359L, 356L, 635L, 615L, 613L, 613L, 617L, 615L, 528L, 476L,
493L, 490L, 492L, 480L)), row.names = c(1L, 2L, 3L, 4L, 5L, 6L,
1002L, 1003L, 1004L, 1005L, 1006L, 1007L, 2003L, 2004L, 2005L,
2006L, 2007L, 2008L, 3004L, 3005L, 3006L, 3007L, 3008L, 3009L
), class = "data.frame")
I would like to run ks test on my different groups.
My groups in the data-frame as such "HINT-binary" "HINT-comp" "InBioMap" "Menche-2015"
Here I found one 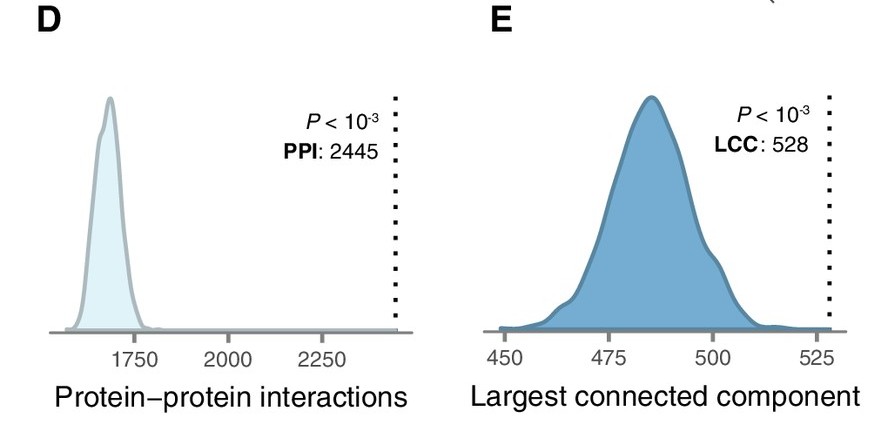 this is what I'm trying to replicate KS test
this is what I'm trying to replicate KS test
The description for the figure give as such
(D) Number of protein-protein interactions (PPIs) between LC genes observed in the high-confidence human interactome (Menche et al., 2015) (dotted line) and 1000 randomized interactome networks (density), revealing significant enrichment for PPIs between LC genes relative to random expectation (p < 10−3). (E) Size of the largest connected component (LCC) between LC genes in the high-confidence human interactome (dotted line) and 1000 randomized interactome networks (density), revealing LC genes occupy a distinct region of the human interactome (p < 10−3). (F) LC genes are prioritized by a disease gene prediction algorithm (Ghiassian et al., 2015) (p < 10−15, Kolmogorov–Smirnov test).
CodePudding user response:
Consider combn to pass pairwise combinations of those groups into ks.test method:
# BUILD NESTED LIST OF RESULTS
ks_results <- combn(
unique(eee$interactome),
2,
FUN = \(x) list(
PPI_ks_results = ks.test(
eee$PPI[eee$interactome == x[1]], eee$PPI[eee$interactome == x[2]]
),
LCC_ks_results = ks.test(
eee$LCC[eee$interactome == x[1]], eee$LCC[eee$interactome == x[2]]
)
),
simplify = FALSE
)
# NAME LIST ELEMENTS
ks_results_names <- setNames(
ks_results,
combn(
unique(eee$interactome), 2, simplify = FALSE
) |> lapply(
\(x) paste(x, collapse="_")
)
)
Output
# REVIEW LIST AND ELEMENTS
str(ks_results)
# List of 6
# $ HINT-binary_HINT-comp :List of 2
# ..$ PPI_ks_results:List of 5
# .. ..$ statistic : Named num 0.833
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.026
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$PPI[eee$interactome == x[1]] and eee$PPI[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# ..$ LCC_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00216
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$LCC[eee$interactome == x[1]] and eee$LCC[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# $ HINT-binary_InBioMap :List of 2
# ..$ PPI_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00216
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$PPI[eee$interactome == x[1]] and eee$PPI[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# ..$ LCC_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00496
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$LCC[eee$interactome == x[1]] and eee$LCC[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# $ HINT-binary_Menche-2015:List of 2
# ..$ PPI_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00216
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$PPI[eee$interactome == x[1]] and eee$PPI[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# ..$ LCC_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00216
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$LCC[eee$interactome == x[1]] and eee$LCC[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# $ HINT-comp_InBioMap :List of 2
# ..$ PPI_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00216
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$PPI[eee$interactome == x[1]] and eee$PPI[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# ..$ LCC_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00496
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$LCC[eee$interactome == x[1]] and eee$LCC[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# $ HINT-comp_Menche-2015 :List of 2
# ..$ PPI_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00216
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$PPI[eee$interactome == x[1]] and eee$PPI[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# ..$ LCC_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00216
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$LCC[eee$interactome == x[1]] and eee$LCC[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# $ InBioMap_Menche-2015 :List of 2
# ..$ PPI_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00216
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$PPI[eee$interactome == x[1]] and eee$PPI[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
# ..$ LCC_ks_results:List of 5
# .. ..$ statistic : Named num 1
# .. .. ..- attr(*, "names")= chr "D"
# .. ..$ p.value : num 0.00496
# .. ..$ alternative: chr "two-sided"
# .. ..$ method : chr "Two-sample Kolmogorov-Smirnov test"
# .. ..$ data.name : chr "eee$LCC[eee$interactome == x[1]] and eee$LCC[eee$interactome == x[2]]"
# .. ..- attr(*, "class")= chr "htest"
Access Individual Elements
ks_results$`HINT-binary_HINT-comp`$PPI_ks_results$statistic
# D
# 0.8333333
ks_results$`HINT-binary_HINT-comp`$PPI_ks_results$p.value
# [1] 0.02597403
Bind to Data Frame
data.frame(
statistic = sapply(ks_results, \(x) x$PPI_ks_results$statistic),
p_value = sapply(ks_results, \(x) x$PPI_ks_results$p.value),
alternative = sapply(ks_results, \(x) x$PPI_ks_results$alternative),
method = sapply(ks_results, \(x) x$PPI_ks_results$method)
)
# statistic p_value alternative method
# HINT-binary_HINT-comp.D 0.8333333 0.025974026 two-sided Two-sample Kolmogorov-Smirnov test
# HINT-binary_InBioMap.D 1.0000000 0.002164502 two-sided Two-sample Kolmogorov-Smirnov test
# HINT-binary_Menche-2015.D 1.0000000 0.002164502 two-sided Two-sample Kolmogorov-Smirnov test
# HINT-comp_InBioMap.D 1.0000000 0.002164502 two-sided Two-sample Kolmogorov-Smirnov test
# HINT-comp_Menche-2015.D 1.0000000 0.002164502 two-sided Two-sample Kolmogorov-Smirnov test
# InBioMap_Menche-2015.D 1.0000000 0.002164502 two-sided Two-sample Kolmogorov-Smirnov test
