So I have the following dataframe:
and what I want is to find the combination of genes that are present together the most.
sample genea geneb genec gened genee genef
1 1 1 1 1 0 0 0
2 2 1 1 1 0 0 0
3 3 1 0 0 1 1 1
4 4 0 0 0 0 0 0
5 5 1 0 1 1 1 1
6 6 0 0 0 0 0 0
so in this case, my desired output would be: gene a c = 3 samples overlap.
test[sort.list(colSums(test[,-1]), decreasing=TRUE)[1:15] 1]) gives me a list with most 1 values per gene. But I am getting stuck with this.
How do I approach this.
CodePudding user response:
One way would be to use crossprod():
library(tidyr)
library(dplyr)
dat %>%
pivot_longer(-sample) %>%
filter(value == 1) %>%
select(-value) %>%
table() %>%
crossprod() %>%
replace(lower.tri(., diag = TRUE), NA) %>%
as.data.frame.table() %>%
slice_max(Freq)
name name.1 Freq
1 genea genec 3
CodePudding user response:
Might not be suitable for every case, but here's a start:
comb <- combn(colnames(df[-1]), m = 2)
overlap <- sapply(1:ncol(comb), \(i) sum(rowSums(df[comb[, i]]) == 2))
list(max.overlap = max(overlap),
wm = df[comb[, which.max(overlap)]])
# $max.overlap
# [1] 3
#
# $wm
# genea genec
# 1 1 1
# 2 1 1
# 3 1 0
# 4 0 0
# 5 1 1
# 6 0 0
CodePudding user response:
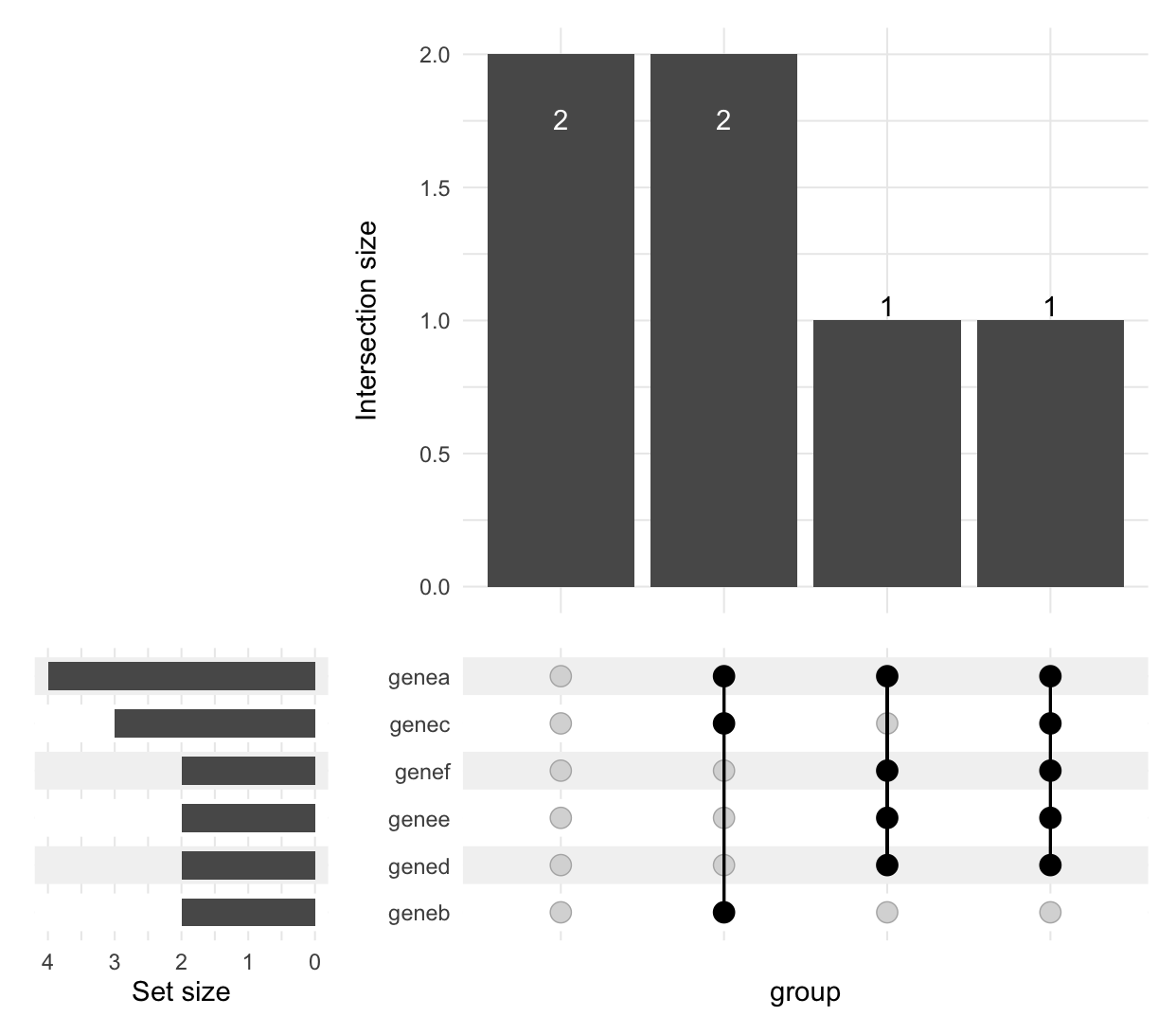
If you want to visualize intersections, you could use ComplexUpset package :
library(ComplexUpset)
ComplexUpset::upset(test[,-1],colnames(test[,-1]))
But it might not be useful if you apply it to a lot of genes.
CodePudding user response:
Data:
test <- data.frame(sample = 1:6,
genea = c(1L, 1L, 1L, 0L, 1L, 0L),
geneb = c(1L, 1L, 0L, 0L, 0L, 0L),
genec = c(1L, 1L, 0L, 0L, 1L, 0L),
gened = c(0L, 0L, 1L, 0L, 1L, 0L),
genee = c(0L, 0L, 1L, 0L, 1L, 0L),
genef = c(0L, 0L, 1L, 0L, 1L, 0L))
Counting the matches:
data.frame(
count = colSums(combn(subset(test,, -sample), 2, rowSums) == 2),
gene = t(combn(subset(test,, -sample), 2, colnames )))
#> count gene.1 gene.2
#> 1 2 genea geneb
#> 2 3 genea genec
#> 3 2 genea gened
#> 4 2 genea genee
#> 5 2 genea genef
#> 6 2 geneb genec
#> 7 0 geneb gened
#> 8 0 geneb genee
#> 9 0 geneb genef
#> 10 1 genec gened
#> 11 1 genec genee
#> 12 1 genec genef
#> 13 2 gened genee
#> 14 2 gened genef
#> 15 2 genee genef
Filtering those with the most matches
subset(
data.frame(
count = colSums(combn(subset(test,, -sample), 2, rowSums) == 2),
gene = t(combn(subset(test,, -sample), 2, colnames ))),
count == max(count))
#> count gene.1 gene.2
#> 2 3 genea genec
Created on 2022-11-09 with reprex v2.0.2