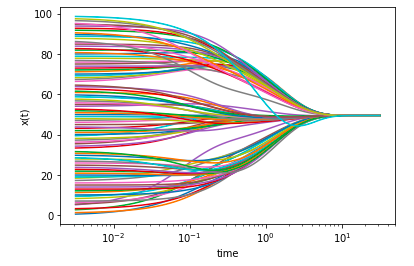
I have a 100-dimensional vector (array) as a result from a function, and I would like to see its change during "time", that is at every step, the elements in my vector assume different values, in a single plot, for every value of the 100 in the vector. What I should get is something like this:
In my context, I have:
def consensus_dynamic(x_0,t):
L=csgraph.laplacian(Last_adj)
x_t=np.dot(linalg.expm(-t*L),x_0)
return x_t
x_t is the 100-dim vector I want to use in the plot, and t the "time". For arbitrary x_0 and t I should obtain that type of graph. I used:
x_0=np.arange(100)
t = np.logspace(-2.5,1.5,100)
Then
x_t=consensus_dynamic(x_0,t)
and with
fig,ax = plt.subplots()
ax.set_xscale('log')
plt.plot(t,x_t)
plt.show()
Can someone help me please?
EDIT: after some tries, I used this code, which computes my vector at every step of t:
steps=[]
for i in t:
x_i=consensus_dynamic(x_0,i)
steps.append(x_i)
plt.plot(t,steps)
plt.show()
I don't know if this is right, but seems closer to the solution than before.
CodePudding user response:
In the end I accomplished it:
t = np.logspace(-2.5,1.5,100)
x_0=np.arange(100)
steps=[]
fig,ax = plt.subplots()
ax.set_xscale('log')
ax.set_xlabel('time')
ax.set_ylabel('x(t)')
for i in t:
x_i=consensus_dynamic(x_0,i)
steps.append(x_i)
plt.plot(t,steps)
plt.show()