I have been working on code that will extract four rows from every .CSV in a folder, compile the rows into a new table, and pivot wider so there is only one entry per each original .csv.
Everything works great until I try to write the resulting table to a .csv,
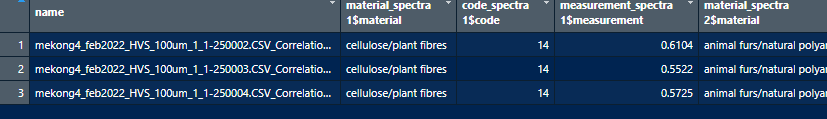
The table looks great in viewer and as a tibble. If I could just get this format to export to .csv I'd be thrilled.
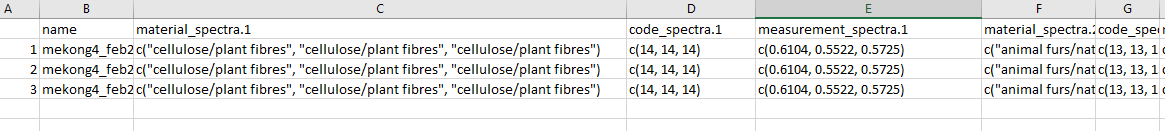
However, when I export it using write.csv I get this:
It looks like each column is being exported as a cell and then copied for each row. I figure the problem is something to do with exporting after completing the pivot_wider function.
Here is my code. Everything works and I am not getting any errors, it just doesn't export the way I need it to.
library(dplyr)
library(tidyverse)
library(tidyr)
#Create a list of files
files <-list.files(pattern = "*Correlations.csv")
# Create an empty object to store all combined rows
combined_df= data.frame(material=character(),
code=double(),
measurement=double(),
name=character(),
spectra=character())
#Create empty object to temporarily collect data produced every run in the loop
data_it= data.frame(matrix(ncol = 5, nrow = 4))
colnames(data_it) =c("material", "code", "measurement", "name", "spectra")
#Run a loop
for (j in 1:length(files)){
data <- read_csv(files[j], skip = 2, col_names =c("material", "code", "measurement", "name", "spectra"))
data_it$material <- data[3:6, 1]
data_it$code <- data[3:6, 2]
data_it$measurement<- data[3:6, 3]
data_it$name <- paste(files[j])
data_it$spectra <- c("spectra 1", "spectra 2", "spectra 3", "spectra 4")
combined_df <- rbind(combined_df, data_it)
}
#pivot table so that there is one row per file
combined_pivot<-pivot_wider(combined_df,
names_from = spectra,
values_from = c(material, code, measurement),
names_vary = "slowest")
#export to .csv
write.csv(combined_pivot, file ="combined_results.csv")
Here is a sample of one of the .csv files I am trying to extract from. I have hundreds of these with 100 rows each and I just need the top three rows from each one
| material | code | measurement |
|---|---|---|
| animal furs/natural polyamides | 13 | 0.6777 |
| animal furs/natural polyamides | 13 | 0.6065 |
| cellulose/plant fibres | 14 | 0.5725 |
| animal furs/natural polyamides | 13 | 0.5698 |
| animal furs/natural polyamides | 13 | 0.5171 |
| animal furs/natural polyamides | 13 | 0.5128 |
| animal furs/natural polyamides | 13 | 0.4932 |
| animal furs/natural polyamides | 13 | 0.4904 |
Thanks for reading and for any suggestions. I am new to R and out of ideas.
CodePudding user response:
Without the data to check and write code, I cannot fix it, however I see the fault. So take this as suggestions rather than answers.
You are generating lists (of 4 things) inside each "cell" of combined_df.
I think you need to pivot_wider inside your for loop before you rbind, so as to break up each files data into 4 lines before you start to add them to a "master" table.
The other way to do it (using for loops) is to have a nested loop inside your original loop that is doing the 3:6 part to a new line each time.
for (i in 3:6){
data_it$material[some variable in here that represents a new line each time] <- data[i, 1]}
as an example, extrapolate from there. The first way seems easier. Good luck
EDIT again:
I mean, you really need a triple nested loop.
One to grab the files
Next to increment your final table each row as you grab the data.
Next to grab the data each line 3:6 (Also has the nested counter to increment the 2nd for loop)
CodePudding user response:
I was able to get it to export by using:
combined_df2 <-as.data.table(combined_df)
before running the pivot.