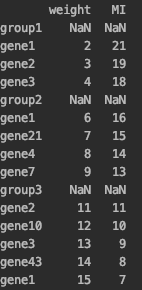
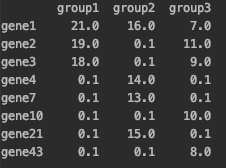
I have a dataset like this
data = {'weight': ['NaN',2,3,4,'NaN',6,7,8,9,'NaN',11,12,13,14,15],
'MI': ['NaN', 21, 19, 18, 'NaN',16,15,14,13,'NaN',11,10,9,8,7]}
df = pd.DataFrame(data, index= ['group1', "gene1", "gene2", 'gene3',
'group2', "gene1", 'gene21', 'gene4', 'gene7', 'group3',
'gene2', 'gene10', 'gene3', 'gene43', 'gene1'])
I need to stack it to gene by group dataframe with MI values. If there are no gene values for particular group the imputated value should be 0.1. 'weights' column should be removed. The final dataframe should look like this
CodePudding user response:
You can use:
m = df['weight'].ne('NaN')
(df[m]
.set_index((~m).cumsum()[m], append=True)['MI']
.unstack('weight', fill_value=0.1)
.add_prefix('group')
)
Variant with pivot:
m = df['weight'].ne('NaN')
(df.assign(col=(~m).cumsum())
.loc[m]
.pivot(columns='col', values='MI')
.fillna(0.1)
.add_prefix('group')
)
Output:
weight group1 group2 group3
gene1 21 16 7
gene10 0.1 0.1 10
gene2 19 0.1 11
gene21 0.1 15 0.1
gene3 18 0.1 9
gene4 0.1 14 0.1
gene43 0.1 0.1 8
gene7 0.1 13 0.1
CodePudding user response:
Another option, still with pivot:
(df
.assign(
grp = np.where(df.weight.eq('NaN'), df.index, np.nan),
group = lambda df: df.grp.ffill())
.loc[lambda df: df.weight.ne('NaN'), ['MI', 'group']]
.pivot(index=None, columns='group', values='MI')
.fillna(0.1)
)
group group1 group2 group3
gene1 21.0 16.0 7.0
gene10 0.1 0.1 10.0
gene2 19.0 0.1 11.0
gene21 0.1 15.0 0.1
gene3 18.0 0.1 9.0
gene4 0.1 14.0 0.1
gene43 0.1 0.1 8.0
gene7 0.1 13.0 0.1
Another process, for the fun of it, and possibly an inkling into the complexities that pivot abstracts:
# get positions for group1,2,3:
positions = df.weight.eq('NaN').to_numpy().nonzero()[0]
groups = df.index[positions]
#get lengths of rows between group1,2,3:
positions = np.diff(np.append(positions, len(df))) - 1
# compute the cumulative positions and pass it to np.split:
arrays = np.split(df.MI.loc[df.MI.ne('NaN')], np.cumsum(positions))
#concatenate the arrays, get the final output:
pd.concat(arrays, axis = 1, keys = groups).fillna(0.1)
group1 group2 group3
gene1 21.0 16.0 7.0
gene2 19.0 0.1 11.0
gene3 18.0 0.1 9.0
gene21 0.1 15.0 0.1
gene4 0.1 14.0 0.1
gene7 0.1 13.0 0.1
gene10 0.1 0.1 10.0
gene43 0.1 0.1 8.0