I have following data:
dput(data)
structure(list(category = c("GO:BP", "GO:BP", "GO:BP", "GO:BP",
"GO:BP", "GO:BP", "GO:BP", "GO:BP", "GO:BP", "GO:BP", "GO:BP",
"GO:BP", "GO:BP", "GO:BP", "GO:BP", "GO:BP", "Pathway", "GO:BP",
"GO:BP", "GO:BP", "GO:BP", "GO:BP", "GO:BP"), ID = c("cell adhesion",
"inflammatory response", "regulation of cell migration", "T cell activation",
"innate immune response", "adaptive immune response", "angiogenesis",
"cytokine-mediated signaling pathway", "wound healing", "regulation of ERK1 and ERK2 cascade",
"extracellular matrix organization", "CD4-positive, alpha-beta T cell activation",
"receptor signaling pathway via JAK-STAT", "natural killer cell activation",
"mast cell activation", "CD8-positive, alpha-beta T cell activation",
"Interleukin-4 and 13 signaling", "regulation of hormone levels",
"response to hypoxia", "regulation of protein tyrosine kinase activity",
"ERBB2-ERBB3 signaling pathway", "fatty acid derivative catabolic process",
"cytolysis"), term = c("GO:0007155", "GO:0006954", "GO:0030334",
"GO:0042110", "GO:0045087", "GO:0002250", "GO:0001525", "GO:0019221",
"GO:0042060", "GO:0070372", "GO:0030198", "GO:0035710", "GO:0007259",
"GO:0030101", "GO:0045576", "GO:0036037", "1470923", "GO:0010817",
"GO:0001666", "GO:0061097", "GO:0038133", "GO:1901569", "GO:0019835"
), Genes = c("ERBB2,ACP5,IKZF1,GRHL2,SIRPG,NLGN4X,CCL4,CCL5,MMP2,CCL19,XCL1,ALOX15,FOXC2,SELE,SELL,NID2,SFRP2,ANK3,EMB,OLFM4,NTNG1,ST3GAL4,NLGN1,CYFIP2,TMEFF1,LY6D,RHOH,SKAP1,SOCS1,MYBPC1,ADAM19,CD244,AXL,LRRC32,TNFRSF14,IL20RB,NFASC,NCAM1,CD84,CCN4,AJAP1,ADAMDEC1,PCDH11X,SYK,NINJ2,GNRH1,LILRB4,LILRA1,LILRB3,PCDHB7,PCDHB4,PCDHA11,SRCIN1,ADAMTS13,RUNX3,CD160,TGM2,CDH22,CD4,CD5,CD6,THY1,TIGIT,CD86,ICAM5,CD37,CD70,CNTN3,TNFAIP3,HAS2,CDH11,FRMD5,CDH15,LILRA4,NRG1,IL32,PDE3B,HLA-DOA,HLA-DQA1,HLA-G,CD83,TNFSF4,ICOS,CCL28,SASH3,PIK3R1,CCR7,PLEK,ONECUT2,ZAN,RHOD,RASAL3,PRKCQ,IL2RG,IL7R,IL12B,RELN,ZBTB16,PSG4,ABCA12,IRF1,SPOCK2,ITGA1,PTAFR,ITGA5,ASTL,ITGB2,ITGB6,LAPTM5,JAK3,PTPRC,PXDN,PTPRJ,CXCR4,NECTIN2,SIGLEC10,TWSG1,DMD,AMTN,VMP1,RAG1,GNE,DSG1,DSG3,CD177,L1CAM,GSC,LIF,CELSR3,EGFR,EBI3,PLXNC1,TENM1,LSAMP,MPZL2,LEF1,EPHB1",
"ACP5,LRRK2,ADCY1,TLR8,MEFV,SIRPG,ADM,CXCL9,CCL4,CCL5,MMP3,CCL18,CCL19,XCL1,ALOX15,SELE,FPR3,BIRC3,MST1,DUOXA1,TRAF3IP2,AXL,C5AR2,IL20RB,CCN4,SYK,LILRB4,CAMK4,SOCS3,MUC19,TF,OSM,CD6,IL1RL1,TNFAIP3,TPSAB1,HGF,HLA-G,CHRNA7,TNFSF4,SERPINA1,CLU,CCR7,ABCC2,DUOX2,NOX4,CCL4L2,CR2,NR1D1,PRKCQ,CYBB,DAB2IP,SLAMF8,IL12B,IRF5,PTAFR,PTGDR,ACE,ITGB2,ITGB6,PTPRC,MYLK3,CXCR4,SIGLEC10,LBP,ECM1,BCL6B,ACE2,NAMPT,LPL,LTA,CALCRL,LYZ",
"ACP5,MTUS1,CCL4,CCL5,MMP1,MMP2,MMP3,CCL18,CCL19,MMP10,VEGFD,XCL1,FOXC2,SELE,SELL,SFRP2,FPR3,OLFM4,NTNG1,ST3GAL4,MST1,RHOH,MAPK15,SOCS1,STK26,TNFRSF14,C5AR2,FGF17,CCN4,KISS1R,GLI1,GLUL,SYK,GNRH1,LDLRAD4,IL24,SASH1,SRCIN1,THY1,PAX6,HAS2,FRMD5,HGF,NRG1,TNFSF4,CCL28,AMOT,PIK3R1,CCR7,ONECUT2,RHOD,NOX4,CCL4L2,RTN4RL2,CTSH,DAB2IP,SLAMF8,RELN,PSG4,PTAFR,ITGA5,ACE,PTPRC,PTPRJ,CXCR4,LBP,ECM1,EGFR,PLXNC1,LGR6,LEF1,FBXW7",
"ERBB2,IKZF1,SIRPG,EOMES,NDRG1,CCL5,CCL19,XCL1,RHOH,SOCS1,TRAF3IP2,CD244,LRRC32,TNFRSF14,IL20RB,CD84,SYK,GNRH1,LILRB4,LILRA1,LILRB3,CAMK4,RUNX3,CD160,CD4,CD5,CD6,CD7,THY1,TIGIT,CD86,TNFSF8,CD37,CD70,LILRA4,HLA-DOA,HLA-DQA1,HLA-G,CD83,CHRNA7,TNFSF4,ICOS,SASH3,PIK3R1,SLC39A6,CCR7,RASAL3,PRKCQ,IL2RG,IL7R,IL12B,TNFRSF9,ZBTB16,PSG4,IRF1,ITGB2,ITK,LAPTM5,JAK3,PTPRC,CXCR4,TWSG1,RAG1,LCP1,LMO1,EBI3,MPZL2,LEF1",
"IFI27L2,LRRK2,TLR8,MEFV,SIRPG,CCL4,CCL5,MMP2,CCL18,CCL19,XCL1,FPR3,BIRC3,APOL1,FASLG,SLA,MST1,MST1R,SLPI,SOCS1,RAET1L,CD244,AXL,TNFRSF14,CD84,SYK,C1QB,C2,C4BPA,C6,KYNU,SOCS3,ADAMTS13,CD160,CD6,CD86,GZMB,CD69,TNFAIP3,LILRA4,HLA-DQA1,HLA-G,SLC30A8,APOBEC3G,TNFSF4,CLU,IRF8,CCL4L2,MED1,CR2,TRAF4,JCHAIN,CYBB,DAB2IP,SLAMF8,IL12B,PSG4,IRF1,IRF5,JAK3,NECTIN2,SIGLEC10,CLNK,KLRD1,CD177,LBP",
"TLR8,EOMES,CCL19,XCL1,ALOX15,SKAP1,TRAF3IP2,CD244,TNFRSF14,IL20RB,SAMSN1,CD84,SYK,C1QB,C2,C4BPA,C6,LILRB4,LILRA1,LILRB3,CAMK4,CD160,CD1B,CD4,CD6,CD7,CD86,GZMB,CD70,IL1RL1,TNFAIP3,LILRA4,HLA-DOA,HLA-DQA1,HLA-G,TNFSF4,CLU,SASH3,CCR7,CR2,JCHAIN,PRKCQ,CTSH,IL7R,IL9R,IL12B,PSG4,IRF1,ITK,JAK3,PTPRC,NECTIN2,SIGLEC10,KLRD1,RAG1,LAIR1,EBI3,LTA,LEF1",
"ERBB2,S100A1,PLEKHH1,ADM,APLNR,CCL5,MMP2,VEGFD,FOXC2,SFRP2,NLGN1,FASLG,ADGRB2,ADGRB3,GJA5,APLN,KLF5,GLUL,SYK,C6,THSD7A,SASH1,CD160,THY1,TNFAIP3,HGF,PDE3B,HLA-G,CHRNA7,HOXB3,AMOT,MED1,CTSH,CYBB,DAB2IP,IL12B,PSG4,ITGA5,ACE,ITGB2,PXDN,CXCR4,TWSG1,ECM1,ADGRG6,LIF,HMGA2,ANGPTL4,VHLL,CALCRL,LEF1,FBXW7,EPHB1",
"CXCL9,CCL4,CCL5,CCL18,CCL19,XCL1,BIRC3,SLC1A1,MST1R,RHEX,SOCS1,TRAF3IP2,AXL,TNFRSF14,IL20RB,SYK,LILRB4,LILRA1,LILRB3,OSM,CD4,CD70,IL1RL1,TNFAIP3,LILRA4,CCR7,CNTFR,CD300LF,DUOX2,CCL4L2,MED1,CSF2RA,IL2RB,IL2RG,IL7R,IL9R,IL10RA,IL12B,PSG4,IRF1,IRF5,LAPTM5,JAK3,PTPRC,PXDN,PTPRJ,CXCR4,CCDC3,ECM1,EBI3",
"ERBB2,F2RL2,MMP3,ALOX15,FOXC2,ST3GAL4,ENPP4,CYP4F11,AXL,SPRR3,LRRC32,CCN4,AJAP1,SYK,LILRB4,IL24,ADAMTS13,TFF3,ENTPD2,TNFAIP3,PAX7,TPSAB1,HGFAC,NRG1,SERPINA1,PLAT,PLEK,POU2F3,DUOX2,PRKCQ,PSG4,DGKG,ITGA5,ITGB6,PTPRJ,CXCR4,TFPI2,LCP1",
"ERBB2,SIRPG,CCL4,CCL5,CCL18,CCL19,XCL1,ALOX15,FPR3,NEK10,C5AR2,SYK,PHLPP1,C1QL4,CD4,NRG1,CHRNA7,CCR7,GAREM1,NOX4,CCL4L2,DAB2IP,PSG4,TBC1D10C,PTPRC,CXCR4,DMD,L1CAM,ACE2,LIF,EGFR,FBXW7,EPHB1",
"MATN2,MATN3,MMP1,MMP2,MMP3,MMP10,FOXF2,FOXC2,NID2,SFRP2,NTNG1,COLGALT2,WDR72,TMPRSS6,ADAMTS13,P4HA1,HAS2,TPSAB1,ERO1B,ADAMTS2,CST2,CST3,SPOCK2,ITGA5,SCX,PXDN,CRISPLD2,LCP1,ADAMTSL3,VHLL",
"CCL19,XCL1,SOCS1,TNFRSF14,LILRB4,RUNX3,CD160,CD86,CD83,TNFSF4,SASH3,CCR7,PRKCQ,IL2RG,IL12B,PSG4,IRF1,JAK3,TWSG1,LEF1",
"CCL5,AKR1B1,MST1,SOCS1,STRA6,IL24,SOCS3,OSM,CSF2RA,IL7R,IL10RA,IL12B,IRF1,JAK3,PTPRC,LIF,HMGA2,VHLL",
"IKZF1,RAET1L,CD244,AXL,LILRB4,CD160,HLA-G,IL2RB,IL12B,IRF1,ITGB2,JAK3,PTPRC,CLNK,KLRD1",
"NDRG1,RHOH,CD84,SYK,LILRB4,NPPC,THY1,CD300LF,PTGDR,NECTIN2,CLNK,LCP2",
"EOMES,XCL1,SOCS1,CD244,LILRB4,RUNX3,TNFSF8,HLA-G,PSG4,IRF1",
"MMP1,MMP2,MMP3,ALOX15,FASLG,SOCS1,SOCS3,OSM,HGF,PIK3R1,IL2RG,IL12B,ITGB2,JAK3,LBP,LIF",
"ADM,CCL5,MME,AKR1B1,DUOXA1,AKR1D1,BCAT2,KISS1R,APLN,GLUL,PCSK1N,TRPA1,PCLO,CHST8,OSM,OXCT1,PCSK6,PAX6,NRG1,PDE3B,SLC30A8,ALDH8A1,CYP2S1,EXOC3L1,BMP8A,ABCC2,HSD17B1,HSD17B2,ERO1B,DUOX2,CPA3,CPE,MED1,NR1D1,CRABP1,RIMS2,RELN,PLB1,ABCA12,CHST9,ACE,KCNB1,DIO2,ACE2,LIF,HMGA2,LEF1",
"ADM,NDRG1,SCN2A,AK4,MMP2,MMP3,VEGFD,ALDOC,MST1,B3GAT1,APLN,CA9,NPPC,SOCS3,NR4A2,TF,TH,CHRNA7,SERPINA1,PLAT,HSD11B2,HILPDA,VLDLR,NOX4,CST2,CST3,PRKCQ,CYBB,ACE,DDIT3,CXCR4,EGLN3,ANGPTL4,VHLL,LTA",
"ERBB2,CCL5,LILRB4,SRCIN1,THY1,NRG1,VLDLR,NRG2,NOX4,RELN,ACE,PTPRC,EGFR,FBXW7",
"ERBB2,NRG1,NRG2", "ACAT1,CYP4F11,OXCT1,ACOT7,HPGD,NUDT7", "GSDMB,C6,GZMB,CCL28,LBP,LYZ"
), pval = c(9.95e-15, 3.17e-06, 0.00126, 1.92e-11, 0.00055, 1.89e-05,
0.00186, 6.06e-08, 0.00191, 0.000176, 0.000644, 5.6e-06, 0.00152,
0.000378, 0.000796, 5.85e-05, 6.42e-05, 0.000359, 0.00163, 0.000192,
0.000607, 0.000177, 0.000885), Zscore = c(4.528095586, 4.330522446,
2.828427125, 5.820855001, 4.677476657, 5.598123172, 1.785687331,
5.374011537, 2.91998558, 1.566698904, 2.556038602, 4.472135955,
3.299831646, 3.356585567, 2.309401077, 2.529822128, 2.5, -0.145864991,
-0.845154255, -0.534522484, -1.732050808, -0.816496581, -0.816496581
), Regulation = c("Activated", "Activated", "Activated", "Activated",
"Activated", "Activated", "Activated", "Activated", "Activated",
"Activated", "Activated", "Activated", "Activated", "Activated",
"Activated", "Activated", "Activated", "Suppressed", "Suppressed",
"Suppressed", "Suppressed", "Suppressed", "Suppressed"), Hit = c(137L,
73L, 72L, 68L, 66L, 59L, 53L, 50L, 38L, 33L, 30L, 20L, 18L, 15L,
12L, 10L, 16L, 47L, 35L, 14L, 3L, 6L, 6L), Count = c(1742L, 1048L,
1249L, 704L, 1088L, 835L, 873L, 548L, 575L, 417L, 394L, 158L,
204L, 136L, 102L, 55L, 114L, 695L, 512L, 114L, 5L, 22L, 29L),
GeneRatio = c(0.078645235, 0.069656489, 0.057646117, 0.096590909,
0.060661765, 0.070658683, 0.060710195, 0.091240876, 0.066086957,
0.079136691, 0.076142132, 0.126582278, 0.088235294, 0.110294118,
0.117647059, 0.181818182, 0.140350877, 0.067625899, 0.068359375,
0.122807018, 0.6, 0.272727273, 0.206896552), adj_pval = c(1.496e-11,
0.0002292, 0.02438, 7.227e-09, 0.0136, 0.0008949, 0.03311,
7.998e-06, 0.03386, 0.005822, 0.01509, 0.0003627, 0.02857,
0.01037, 0.01746, 0.002417, 0.01402, 0.00996, 0.03023, 0.006202,
0.01458, 0.005828, 0.01901)), class = "data.frame", row.names = c(NA,
-23L))
I used following code to generate the plot:
p <- ggplot(data, aes(Zscore, ID))
p geom_point(aes(colour=pval, size=Hit))
scale_color_gradientn(colours=rainbow(4), limits=c(0, 1))
geom_vline(xintercept=0, size=0.2, colour="blue", linetype="dotted")
theme(panel.background=element_rect(fill="gray95", colour="gray95"),
panel.grid.major=element_line(size=0.1,linetype='solid', colour="gray90"),
panel.grid.minor=element_line(size=0.1,linetype='solid', colour="gray90"),
axis.title.y=element_blank())
expand_limits(x=c(-2,3))
scale_x_continuous(breaks=c(-3,-2,-1,0,1,2,3))
scale_y_discrete(limits=rev(data$ID))
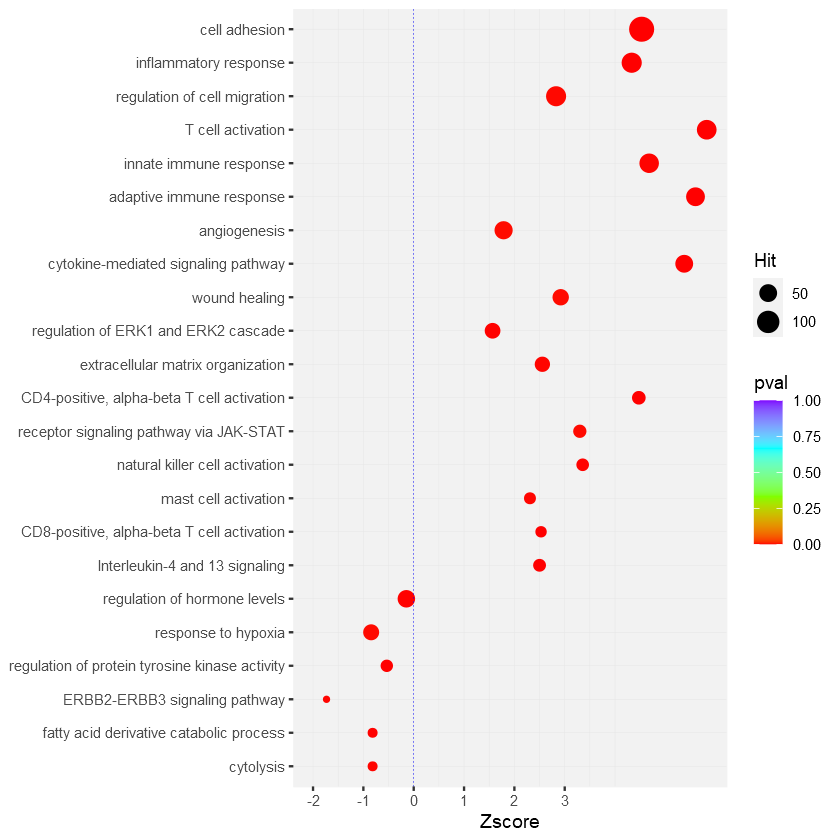 in the output, I want to change the color (to green) of
in the output, I want to change the color (to green) of positive Zscore values. Also to increase the hit to 0,10,20,25,50....
CodePudding user response:
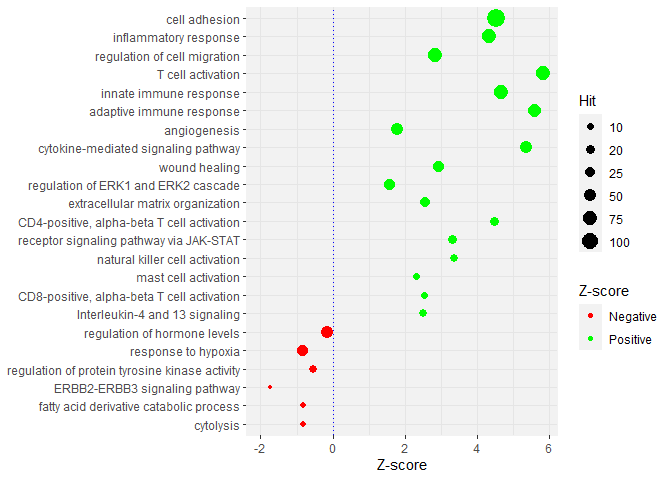
The plot below is maybe not what is wanted.
- To have the color change to green when the Z-score is positive means it should be mapped to
Zscore, not topval; - Also, the aesthetic
sizefor lines was deprecated inggplot2 3.4.0, I am usinglinewidthinstead.
library(ggplot2)
p <- ggplot(data, aes(Zscore, ID))
p geom_point(aes(colour = Zscore > 0, size = Hit))
scale_color_manual(
name = 'Z-score',
labels = c("Negative", "Positive"),
values = c(`FALSE` = 'red', `TRUE` = 'green')
)
scale_size_continuous(breaks = c(0,10,20,25,50, 75, 100))
geom_vline(xintercept=0, linewidth=0.2, colour="blue", linetype="dotted")
theme(panel.background=element_rect(fill="gray95", colour="gray95"),
panel.grid.major=element_line(linewidth=0.1,linetype='solid', colour="gray90"),
panel.grid.minor=element_line(linewidth=0.1,linetype='solid', colour="gray90"),
axis.title.y=element_blank())
expand_limits(x=c(-2,3))
# scale_x_continuous(breaks=c(-3,-2,-1,0,1,2,3, 4, 5, 6))
scale_x_continuous(name = 'Z-score', breaks = pretty(data$Zscore))
scale_y_discrete(limits=rev(data$ID))
Created on 2022-11-18 with reprex v2.0.2
