I have a large dataset in which I want to group similar resistance patterns together. A plot to visualize similarity of resistance pattern is needed.
dat <- read.table(text="Id Resistance.Pattern
A SSRRSSSSR
B SSSRSSSSR
C RRRRSSRRR
D SSSSSSSSS
E SSRSSSSSR
F SSSRRSSRR
G SSSSR
H SSSSSSRRR
I RRSSRRRSS", header=TRUE)
CodePudding user response:
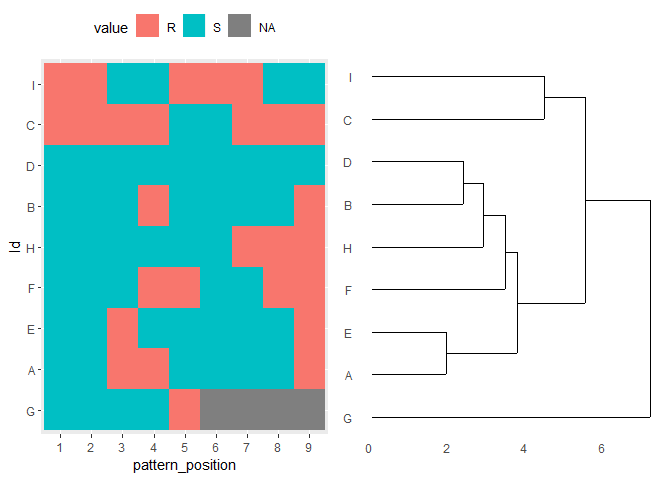
I would separate out the values into a wider dataframe and then make a heatmap and dendrogram to compare sillimanites in patterns:
library(tidyverse)
library(ggdendro)
recode_dat <- dat |>
mutate(pat = str_split(Resistance.Pattern, "")) |>
unnest_wider(pat, names_sep = "_") |>
select(starts_with("pat_")) |>
mutate(across(everything(), ~case_when(. == "S" ~ 1, . == "R" ~ 2, is.na(.) ~0)))
rownames(recode_dat) <- dat$Id
dendro <- as.dendrogram(hclust(d = dist(x = scale(recode_dat))))
dendro_plot <- ggdendrogram(data = dendro, rotate = TRUE)
heatmap_plot <- dat |>
mutate(pat = str_split(Resistance.Pattern, "")) |>
unnest_wider(pat, names_sep = "_") |>
pivot_longer(cols = starts_with("pat_"), names_to = "pattern_position") |>
mutate(Id = factor(Id, levels = dat$Id[order.dendrogram(dendro)])) |>
ggplot(aes(pattern_position, Id))
geom_tile(aes(fill = value))
scale_x_discrete(labels = \(x) sub(".*_(\\d $)", "\\1", x))
theme(legend.position = "top")
cowplot::plot_grid(heatmap_plot, dendro_plot,nrow = 1, align = "h", axis = "tb")
CodePudding user response:
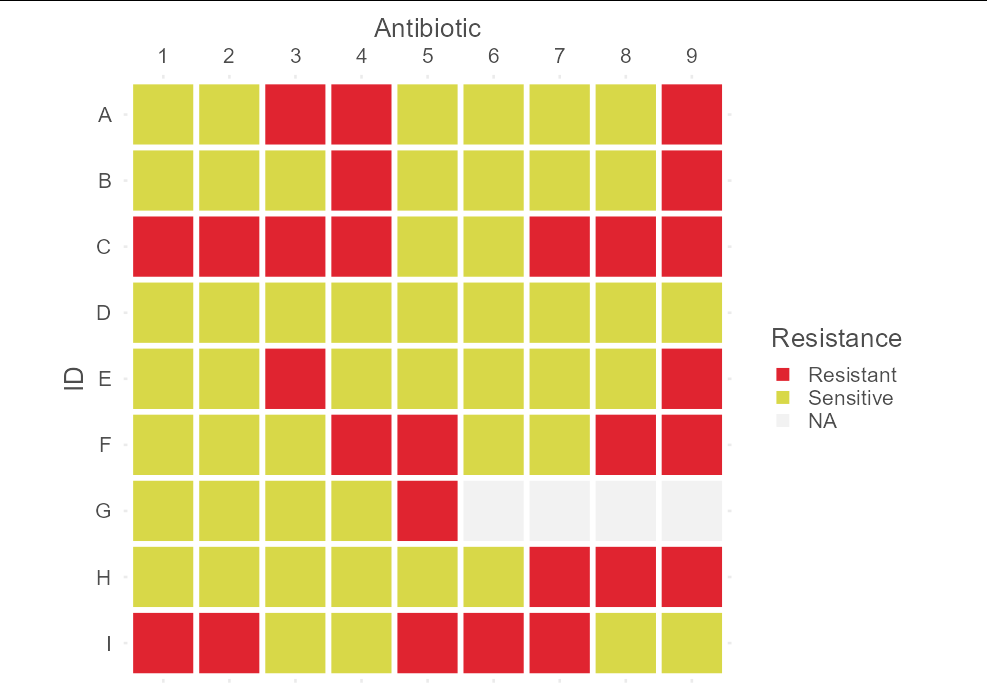
It sounds as though the second column of your data frame represents sensitivity (S) and resistance (R), presumably to antibiotics (though this is not clear in your question). That being the case, you are presumably looking for something like this:
library(tidyverse)
p <- strsplit(dat$Resistance.Pattern, "")
do.call(rbind, lapply(p, \(x) c(x, rep(NA, max(lengths(p)) - length(x))))) %>%
as.data.frame() %>%
cbind(Id = dat$Id) %>%
mutate(Id = factor(Id, rev(Id))) %>%
pivot_longer(V1:V9) %>%
ggplot(aes(name, Id, fill = value))
geom_tile(col = "white", size = 2)
coord_equal()
scale_fill_manual(values = c("#e02430", "#d8d848"),
labels = c("Resistant", "Sensitive"),
na.value = "gray95")
scale_x_discrete(name = "Antibiotic", position = "top",
labels = 1:9)
labs(fill = "Resistance", y = "ID")
theme_minimal(base_size = 20)
theme(text = element_text(color = "gray30"))