I have 2 numpy arrays:
import numpy as np
a = np.array([1, 2, 3])
b = np.array([10, 20, 30])
I need to create a list of dicts:
res =
[{"a": 1, "b": 10},
{"a": 2, "b": 20},
{"a": 3, "b": 30}]
in most optimal way, without iterating through the whole array.
The obvious solution
res = [{"a": a_el, "b": b_el} for a_el, b_el in zip(a, b)]
takes too much time if a and b has a lot of values inside
CodePudding user response:
If you're open to also importing pandas, you could do:
import pandas as pd
df = pd.DataFrame({"a": a, "b": b})
res = df.to_dict(orient='records')
which gives the desired res:
[{'a': 1, 'b': 10}, {'a': 2, 'b': 20}, {'a': 3, 'b': 30}]
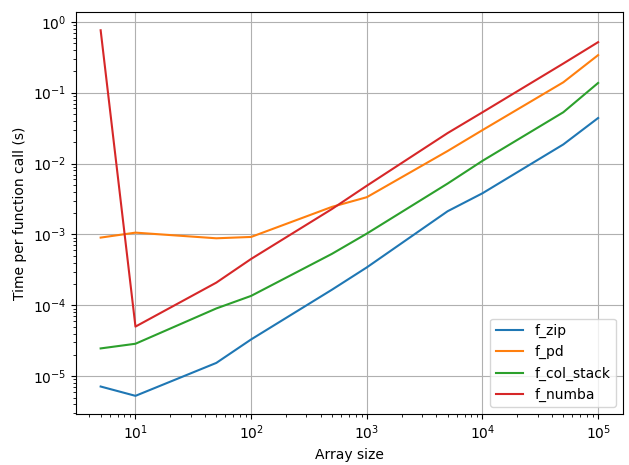
Depending on the size of your arrays, this might not be worth it. It appears this isn't worth it regardless of the size of your arrays, but I'm going to keep this answer for its educational value, and will update it to compare the runtimes of methods that other people suggest.
Timing both approaches, my computer shows the zip approach is always faster than the pandas approach, so disregard the previous part of this answer.
Ranking (fastest to slowest)
- Plain old
zip - 0x0fba's
np.col_stack - My approach -- create a dataframe and
df.to_dict
= crashMOGWAI's numba approach (time for first function call is skewed by compilation time)
import timeit
from numba import jit
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
def f_zip(a, b):
return [{"a": ai, "b": bi} for ai, bi in zip(a, b)]
def f_pd(a, b):
df = pd.DataFrame({"a": a, "b": b})
return df.to_dict(orient='records')
def f_col_stack(a, b):
return [{"a": a, "b": b} for a, b in np.column_stack((a,b))]
@jit
def f_numba(a, b):
return [{"a": a_el, "b": b_el} for a_el, b_el in zip(a, b)]
funcs = [f_zip, f_pd, f_col_stack, f_numba]
sizes = [5, 10, 50, 100, 500, 1000, 5000, 10_000, 50_000, 100_000]
times = np.zeros((len(sizes), len(funcs)))
N = 20
for i, s in enumerate(sizes):
a = np.random.random((s,))
b = np.random.random((s,))
for j, f in enumerate(funcs):
times[i, j] = timeit.timeit("f(a, b)", globals=globals(), number=N) / N
print(".", end="")
print(s)
fig, ax = plt.subplots()
for j, f in enumerate(funcs):
ax.plot(sizes, times[:, j], label=f.__name__)
ax.set_xlabel("Array size")
ax.set_ylabel("Time per function call (s)")
ax.set_xscale("log")
ax.set_yscale("log")
ax.legend()
ax.grid()
fig.tight_layout()
CodePudding user response:
You can try this one-liner that uses a list-comprehension to build the list of dicts, as well as the numpy column_stack() method.
res = [{"a": a, "b": b} for a, b in np.column_stack((a,b))]
CodePudding user response:
I would say your current approach is fairly efficient. Not knowing any other details, you may be able to precompile w/ numba and save a little execution time. Making some order of magnitude and memory availability assumptions, see below Jupyter execution.
# %%
import numpy as np
from numba import jit
# %%
%%time
x = np.array(range(1,1000000,1))
y = np.array(range(10,1000000,10))
test = [{"a": a_el, "b": b_el} for a_el, b_el in zip(x, y)]
# CPU times: total: 234 ms
# Wall time: 231 ms
# %%
%%time
@jit
def f():
a = np.array(range(1,1000000,1))
b = np.array(range(10,1000000,10))
return [{"a": a_el, "b": b_el} for a_el, b_el in zip(a, b)]
# CPU times: total: 46.9 ms
# Wall time: 46.3 ms