I have data that is merged down but on every hour mark, there is a row in my data with an elevation value and a row in my data with a NA. I want to be able to just merge those rows together, or in other words, take out the row that has the NA value. It's okay that all of the other values in the table are NA.
Here's my data:
> dput(stackoverflow[1:100,])
structure(list(Time = structure(c(1432425600, 1432426500, 1432427400,
1432428300, 1432429200, 1432430100, 1432431000, 1432431900, 1432432800,
1432433700, 1432434600, 1432435500, 1432436400, 1432437300, 1432438200,
1432439100, 1432440000, 1432440900, 1432441800, 1432442700, 1432443600,
1432444500, 1432445400, 1432446300, 1432447200, 1432448100, 1432449000,
1432449900, 1432450800, 1432450800, 1432451700, 1432452600, 1432453500,
1432454400, 1432454400, 1432455300, 1432456200, 1432457100, 1432458000,
1432458000, 1432458900, 1432459800, 1432460700, 1432461600, 1432461600,
1432462500, 1432463400, 1432464300, 1432465200, 1432465200, 1432466100,
1432467000, 1432467900, 1432468800, 1432468800, 1432469700, 1432470600,
1432471500, 1432472400, 1432472400, 1432473300, 1432474200, 1432475100,
1432476000, 1432476000, 1432476900, 1432477800, 1432478700, 1432479600,
1432479600, 1432480500, 1432481400, 1432482300, 1432483200, 1432483200,
1432484100, 1432485000, 1432485900, 1432486800, 1432486800, 1432487700,
1432488600, 1432489500, 1432490400, 1432490400, 1432491300, 1432492200,
1432493100, 1432494000, 1432494000, 1432494900, 1432495800, 1432496700,
1432497600, 1432498500, 1432499400, 1432500300, 1432501200, 1432502100,
1432503000), tzone = "UTC", class = c("POSIXct", "POSIXt")),
Turtle = structure(c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L), levels = c("R3L1", "R3L11", "R3L12", "R3L2", "R3L4",
"R3L8", "R3L9", "R4L8", "R8L1", "R8L4", "R8NAT123"), class = "factor"),
elevation = c(NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,
NA, NA, 282.27, NA, NA, NA, NA, 283.21, NA, NA, NA, NA, 282.14,
NA, NA, NA, NA, 281.63, NA, NA, NA, NA, 281.63, NA, NA, NA,
NA, 281.63, NA, NA, NA, NA, 282.63, NA, NA, NA, NA, 281.63,
NA, NA, NA, NA, 282.14, NA, NA, NA, NA, 281.63, NA, NA, NA,
NA, 282.14, NA, NA, NA, NA, 281.36, NA, NA, NA, NA, 282.14,
NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA)), row.names = c(NA,
-100L), class = c("tbl_df", "tbl", "data.frame"))
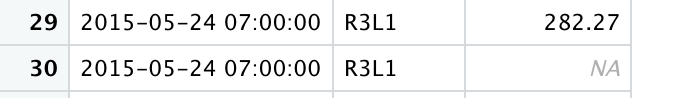 Here's also a picture of an example of rows I would like to merge.
Here's also a picture of an example of rows I would like to merge.
Let me know if you have any questions.
CodePudding user response:
Since the data frame (here, df) has the same Time and Turtle values and the NA values are the only ones repeating and always come after the Elevation value, a dplyr approach would be:
library(dplyr)
df %>%
group_by(Time, Turtle) %>%
slice(1)
# or
df %>% # thanks @neilfws
group_by(Time, Turtle) %>%
slice_head()
Which groups the Time and Turtle values and takes the first value.
CodePudding user response:
with dplyr
library(dplyr)
df1 %>%
group_by(Time, Turtle) %>%
summarise(elevation = elevation[!is.na(elevation)][1], .groups = 'drop')
-output
# A tibble: 87 × 3
# Groups: Time [87]
Time Turtle elevation
<dttm> <fct> <dbl>
1 2015-05-24 00:00:00 R3L1 NA
2 2015-05-24 00:15:00 R3L1 NA
3 2015-05-24 00:30:00 R3L1 NA
4 2015-05-24 00:45:00 R3L1 NA
5 2015-05-24 01:00:00 R3L1 NA
6 2015-05-24 01:15:00 R3L1 NA
7 2015-05-24 01:30:00 R3L1 NA
8 2015-05-24 01:45:00 R3L1 NA
9 2015-05-24 02:00:00 R3L1 NA
10 2015-05-24 02:15:00 R3L1 NA
Or use distinct
df1 %>%
arrange(Time, Turtle, is.na(elevation)) %>%
distinct()
