I am using the cutpointr package to generate cut off for a continuous variable. I work as prescribed but the plot objects generated are complex and a result of large gtable data. I want to format or edit the legend in the plot but I have failed totally with ggplot2
This is the code with cutpointr used to generate the cut off:
opt_cut_b_cycle.type<- cutpointr(hcgdf_v2, beta.hcg, livebirth.factor, cycle.type,
method = maximize_boot_metric,
metric = youden, boot_runs = 1000,
boot_stratify = TRUE,
na.rm = TRUE) %>% add_metric(list(ppv, npv, odds_ratio, risk_ratio, p_chisquared))
The plot object is obtained by running 'plot' function
plot(opt_cut_b_cycle.type)
This is the plot generated
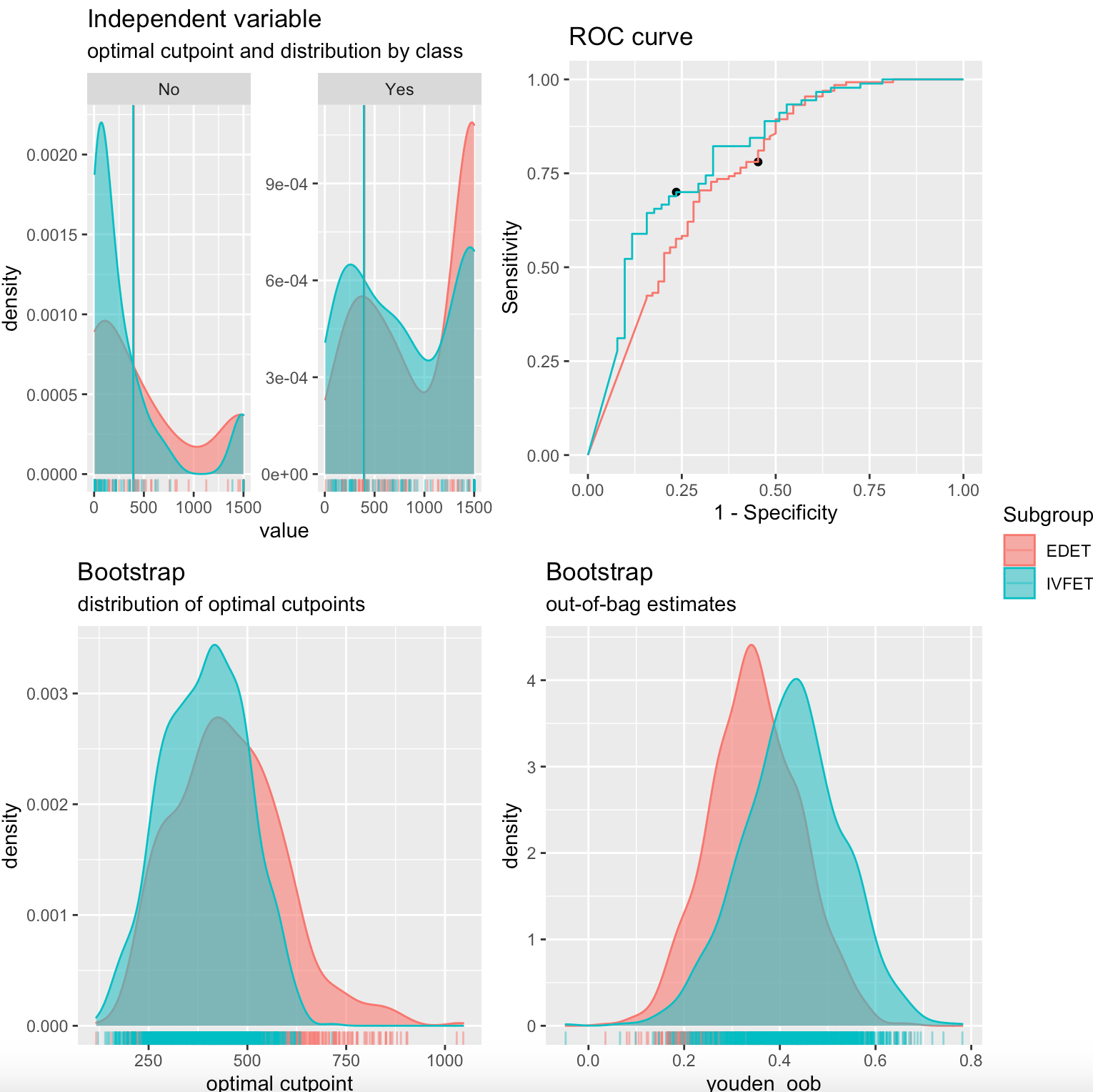 .
.
- I want to edit the legend title from subgroup to Oocyte source
- I want to change the labels EDET to Donor, IVFET to Autologous
I tried working treating the plot object as a ggplot2 plot and running code such as, where p is the said plot object.
p scale_fill_discrete(name = "Oocyte source", labels = c("Donor", "Autologous"))
Unfortunately, the console returns 'NULL'
This is an example data set:
hcgdf_v2 <-tibble(id = 1:10, beta.hcg = seq(from = 5, to = 1500, length.out = 10),
livebirth.factor = c("yes", "no", "yes", "no", "no", "yes", "yes", "no", "no", "yes"),
cycle.type = c("edet","ivfet","edet", "edet", "edet", "edet", "ivfet", "ivfet", "ivfet","edet"))
CodePudding user response:
When I attempted to use your code, it didn't work. However, based on the current type of graph and graph options you've called, this should work.
The legend title
I suggest you run this line and ensure it returns Subgroup before using it to change anything.
# assign the plot to an object
plt <- plot(opt_cut_b_cycle.type)
# printing this should return "Subgroup" - current legend title
plt$grobs[[2]]$grobs[[1]]$grobs[[2]][[4]][[1]][[6]][[1]][[1]]
# change the legend title
plt$grobs[[2]]$grobs[[1]]$grobs[[2]][[4]][[1]][[6]][[1]][[1]] <- "Oocyte"
Legend entries
The easiest method to change the legend entries is probably to rename the factors in your data. Alternatively, you can change these labels the same way you changed the legend title.
Note that the colors will swap between the two options when you change the factor levels. (That's because it is alphabetized.)
#### Option 1 - - Recommended method
# change the legend entries-- factor levels
# in your image, you have "EDET", but your data has "edet"
# make sure this has the capitalization used in your data
hcgdf_v2$cycle2 <- ifelse(hcgdf_v2$cycletype == "edet", "Donor", "Autologous")
# now rerun plot with alternate subgroup
opt_cut_b_cycle.type<- cutpointr(hcgdf_v2, beta.hcg, livebirth.factor, cycle2,
method = maximize_boot_metric,
metric = youden, boot_runs = 1000,
boot_stratify = TRUE,
na.rm = TRUE) %>%
add_metric(list(ppv, npv, odds_ratio, risk_ratio, p_chisquared))
#### Option 2 - - Not recommended due to legend spacing
# alternative to rename legend entry labels
# this should return "EDET"
plt$grobs[[2]]$grobs[[1]]$grobs[[7]][[4]][[1]][[6]][[1]][[1]]
# this should return "IVFET"
plt$grobs[[2]]$grobs[[1]]$grobs[[8]][[4]][[1]][[6]][[1]][[1]]
plt$grobs[[2]]$grobs[[1]]$grobs[[7]][[4]][[1]][[6]][[1]][[1]] <- "Donor"
plt$grobs[[2]]$grobs[[1]]$grobs[[8]][[4]][[1]][[6]][[1]][[1]] <- "Autologous"
To see your modified plot, use plot.
plot(plt)
When I ran it this code, changing the legend title causes some odd behavior where the plot background isn't entirely white, if that happens in your plot do the following.
This requires the library gridExtra.
# clear the plot
plot.new()
# recreate the grid
plt2 <- grid.arrange(plt$grobs[[1]]$grobs[[1]], # 2 small graphs top left
plt$grobs[[1]]$grobs[[2]], # ROC curve graph (top right)
plt$grobs[[1]]$grobs[[3]], # distro of optimal cut
plt$grobs[[1]]$grobs[[4]], nrow = 2) # out-of-bag estimates
plot.new()
# graphs and legend set to 4:1 ratio of space graphs to legend
grid.arrange(plt2, plt$grobs[[2]], ncol = 2, widths = c(4, 1))
```
