I have a dataset (data.weather) with one weather variable (TMAX) for two locations (combination of LAT and LON) and two years. TMAX is available for ten days per year and location in this mock example. I need to calculate the mean TMAX (mean_TMAX) for each of the four rows in data.locs. This last dataset indicate the range of date for which I need to calculate the mean. That is between DATE_0 and DATE_1.
Here is the code of what I did:
library(dplyr)
library(lubridate)
data.weather <-read.csv(text = "
LAT,LON,YEAR,DATE,TMAX
36,-89,2010,1/1/2010,25
36,-89,2010,1/2/2010,25
36,-89,2010,1/3/2010,25
36,-89,2010,1/4/2010,28
36,-89,2010,1/5/2010,28
36,-89,2010,1/6/2010,29
36,-89,2010,1/7/2010,25
36,-89,2010,1/8/2010,25
36,-89,2010,1/9/2010,25
36,-89,2010,1/10/2010,28
36,-89,2011,1/1/2011,26
36,-89,2011,1/2/2011,25
36,-89,2011,1/3/2011,28
36,-89,2011,1/4/2011,26
36,-89,2011,1/5/2011,27
36,-89,2011,1/6/2011,27
36,-89,2011,1/7/2011,28
36,-89,2011,1/8/2011,29
36,-89,2011,1/9/2011,27
36,-89,2011,1/10/2011,26
40,-96,2010,1/1/2010,29
40,-96,2010,1/2/2010,28
40,-96,2010,1/3/2010,25
40,-96,2010,1/4/2010,25
40,-96,2010,1/5/2010,28
40,-96,2010,1/6/2010,29
40,-96,2010,1/7/2010,26
40,-96,2010,1/8/2010,28
40,-96,2010,1/9/2010,26
40,-96,2010,1/10/2010,25
40,-96,2011,1/1/2011,29
40,-96,2011,1/2/2011,27
40,-96,2011,1/3/2011,29
40,-96,2011,1/4/2011,25
40,-96,2011,1/5/2011,28
40,-96,2011,1/6/2011,29
40,-96,2011,1/7/2011,29
40,-96,2011,1/8/2011,25
40,-96,2011,1/9/2011,25
40,-96,2011,1/10/2011,26
") %>%
mutate(DATE = as.Date(DATE, format = "%m/%d/%Y"))
data.locs <-read.csv(text = "
LAT,LON,YEAR,DATE_0,DATE_1,GEN,PR
36,-89,2010,1/2/2010,1/9/2010,MN103,35
36,-89,2011,1/1/2011,1/10/2011,IA100,33
40,-96,2010,1/4/2010,1/8/2010,MN103,36
40,-96,2011,1/2/2011,1/6/2011,IA100,34
") %>%
mutate(DATE_0 = as.Date(DATE_0, format = "%m/%d/%Y"),
DATE_1 = as.Date(DATE_1, format = "%m/%d/%Y"))
tmax.calculation <- data.locs %>%
group_by(LAT,LON,YEAR, GEN) %>%
mutate(mean_TMAX = mean(data.weather$TMAX[data.weather$DATE %within% interval(DATE_0, DATE_1)]))
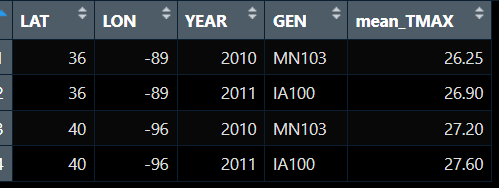
This is the expected result:
LAT LON YEAR DATE_0 DATE_1 GEN PR meam_tmax
36 -89 2010 1/2/2010 1/9/2010 MN103 35 26.25
36 -89 2011 1/1/2011 1/10/2011 IA100 33 26.90
40 -96 2010 1/4/2010 1/8/2010 MN103 36 27.20
40 -96 2011 1/2/2011 1/6/2011 IA100 34 27.60
However, this is what I am getting:
LAT LON YEAR DATE_0 DATE_1 GEN PR meam_tmax
36 -89 2010 1/2/2010 1/9/2010 MN103 35 26.5625
36 -89 2011 1/1/2011 1/10/2011 IA100 33 27.0500
40 -96 2010 1/4/2010 1/8/2010 MN103 36 27.1000
40 -96 2011 1/2/2011 1/6/2011 IA100 34 27.1000
The problem I have is that, when reading the data interval in data.weather, the calculation is being made over the correct interval BUT across the two locations (combination of LAT and LON). I couldn't find a way to indicate to calculate the mean only for each LAT and LON combination separately.
CodePudding user response:
This should do it:
library(dplyr)
library(lubridate)
data.weather <-read.csv(text = "
LAT,LON,YEAR,DATE,TMAX
36,-89,2010,1/1/2010,25
36,-89,2010,1/2/2010,25
36,-89,2010,1/3/2010,25
36,-89,2010,1/4/2010,28
36,-89,2010,1/5/2010,28
36,-89,2010,1/6/2010,29
36,-89,2010,1/7/2010,25
36,-89,2010,1/8/2010,25
36,-89,2010,1/9/2010,25
36,-89,2010,1/10/2010,28
36,-89,2011,1/1/2011,26
36,-89,2011,1/2/2011,25
36,-89,2011,1/3/2011,28
36,-89,2011,1/4/2011,26
36,-89,2011,1/5/2011,27
36,-89,2011,1/6/2011,27
36,-89,2011,1/7/2011,28
36,-89,2011,1/8/2011,29
36,-89,2011,1/9/2011,27
36,-89,2011,1/10/2011,26
40,-96,2010,1/1/2010,29
40,-96,2010,1/2/2010,28
40,-96,2010,1/3/2010,25
40,-96,2010,1/4/2010,25
40,-96,2010,1/5/2010,28
40,-96,2010,1/6/2010,29
40,-96,2010,1/7/2010,26
40,-96,2010,1/8/2010,28
40,-96,2010,1/9/2010,26
40,-96,2010,1/10/2010,25
40,-96,2011,1/1/2011,29
40,-96,2011,1/2/2011,27
40,-96,2011,1/3/2011,29
40,-96,2011,1/4/2011,25
40,-96,2011,1/5/2011,28
40,-96,2011,1/6/2011,29
40,-96,2011,1/7/2011,29
40,-96,2011,1/8/2011,25
40,-96,2011,1/9/2011,25
40,-96,2011,1/10/2011,26
") %>%
mutate(DATE = as.Date(DATE, format = "%m/%d/%Y"))
data.locs <-read.csv(text = "
LAT,LON,YEAR,DATE_0,DATE_1,GEN,PR
36,-89,2010,1/2/2010,1/9/2010,MN103,35
36,-89,2011,1/1/2011,1/10/2011,IA100,33
40,-96,2010,1/4/2010,1/8/2010,MN103,36
40,-96,2011,1/2/2011,1/6/2011,IA100,34
") %>%
mutate(DATE_0 = as.Date(DATE_0, format = "%m/%d/%Y"),
DATE_1 = as.Date(DATE_1, format = "%m/%d/%Y"))
tmax.calculation <- data.locs %>%
group_by(LAT,LON,YEAR,GEN) %>%
do(data.frame(LAT=.$LAT,
LON=.$LON,
YEAR=.$YEAR,
GEN=.$GEN,
DATE=seq(.$DATE_0, .$DATE_1, by="days"))) %>%
left_join(data.weather, by=c("LAT", "LON", "YEAR", "DATE")) %>%
summarise(mean_TMAX = mean(TMAX))
Result: