I have a dataset with 5 levels for treatment and 4 levels for time, and values for germination for those 20 combinations. I am trying to make a grouped bar chart that will show fitted values and confidence intervals from a glm that I have made.
mod_time <- glm(cbind(germination, total - germination) ~ time, data = df, family = binomial)
df$time_fit <- predict(mod_time, newdata = df, interval = "confidence")
conf_int <- confint(mod_time)
ggplot(df, aes(x = treatment, y = germination, fill = time))
geom_col(position = "dodge")
geom_line(aes(y = time_fit), position = position_dodge(width = 0.9), color = "blue")
geom_errorbar(aes(ymin = conf_int[,2], ymax = conf_int[,3]), position = position_dodge(width = 0.9))
labs(x = "Scarification", y = "Germination/Total", fill = "Time")
However this just gives me a grouped bar chart without the confidence intervals and runs up errors about the confidence intervals not having the correct dimensions. The confidence intervals seem to be made into a vector rather than a matrix. Any ideas as to what I am doing wrong?
CodePudding user response:
I didn't have your data, so with some toy data this is how I'd approach this:
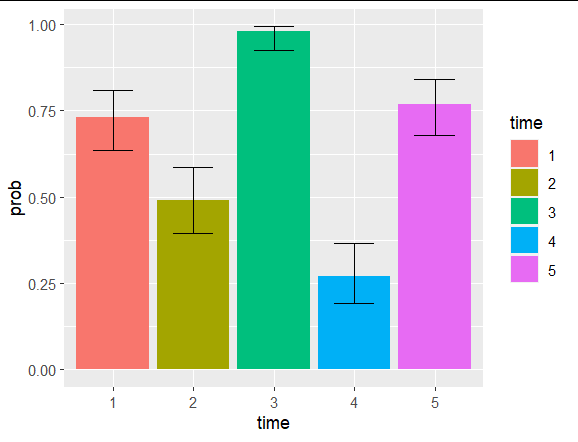
It doesn't look like predict.glm supplies confidence intervals, even with interval="confidence" so I used emmeans to generate a dataset with the mean probabilties and confidence intervals for each group. type=response made sure the results came back on the probability scale not the 'xb' scale.
These are asymptotic CIs so for small counts I would probably not use this method but generate exact CIs instead.
library(ggplot2)
library(emmeans)
dat <- data.frame(time = factor(1:5), germination=sample(100,5), total=100)
mod_time <- glm(cbind(germination, total-germination) ~ time, data=dat, family="binomial")
preds <- as.data.frame(emmeans(mod_time, ~time, type="response"))
ggplot(preds, aes(x=time, y=prob, ymin=asymp.LCL, ymax=asymp.UCL))
geom_col(aes(fill=time))
geom_errorbar(width=0.5)