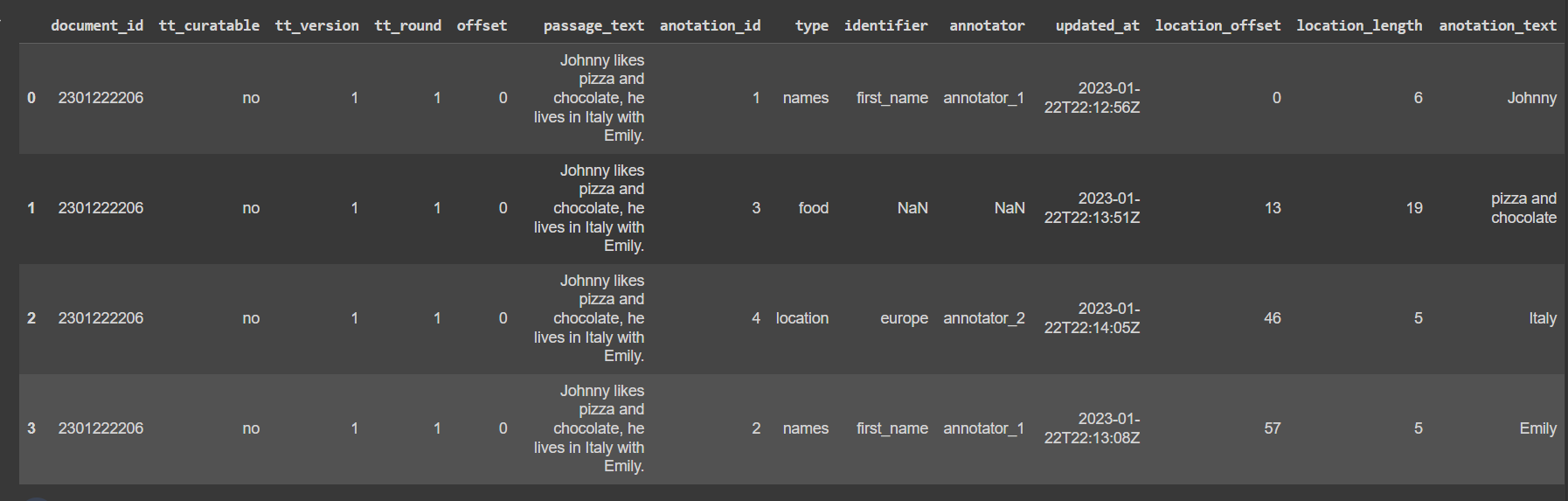
I have a heavily nested xml that I'm trying to convert to a data frame object.
attached failed attempts bellow.
input : johnny.xml file, contains the following text-
<?xml version="1.0" encoding="UTF-8"?>
<!DOCTYPE collection SYSTEM "BioC.dtd">
<collection>
<source/>
<date/>
<key/>
<document>
<id>2301222206</id>
<infon key="tt_curatable">no</infon>
<infon key="tt_version">1</infon>
<infon key="tt_round">1</infon>
<passage>
<offset>0</offset>
<text>Johnny likes pizza and chocolate, he lives in Italy with Emily.</text>
<annotation id="1">
<infon key="type">names</infon>
<infon key="identifier">first_name</infon>
<infon key="annotator">annotator_1</infon>
<infon key="updated_at">2023-01-22T22:12:56Z</infon>
<location offset="0" length="6"/>
<text>Johnny</text>
</annotation>
<annotation id="3">
<infon key="type">food</infon>
<infon key="identifier"></infon>
<infon key="annotator">annotator_2</infon>
<infon key="updated_at">2023-01-22T22:13:51Z</infon>
<location offset="13" length="19"/>
<text>pizza and chocolate</text>
</annotation>
<annotation id="4">
<infon key="type">location</infon>
<infon key="identifier">europe</infon>
<infon key="annotator">annotator_2</infon>
<infon key="updated_at">2023-01-22T22:14:05Z</infon>
<location offset="46" length="5"/>
<text>Italy</text>
</annotation>
<annotation id="2">
<infon key="type">names</infon>
<infon key="identifier">first_name</infon>
<infon key="annotator">annotator_1</infon>
<infon key="updated_at">2023-01-22T22:13:08Z</infon>
<location offset="57" length="5"/>
<text>Emily</text>
</annotation>
</passage>
</document>
</collection>
failed attempts:
- with lxml
from lxml import objectify
root = objectify.parse('johnny.xml').getroot()
data=[]
for i in range(len(root.getchildren())):
data.append([child.text for child in root.getchildren()[i].getchildren()])
df = pd.DataFrame(data)
result -
0 1 2 3 4
0 None None None None None
1 None None None None None
2 None None None None None
3 2301222206 no 1 1 None
- this solution using recursive function (2nd answer) result -
id infon-1 infon-2 infon-3 infon-key-1 infon-key-2 infon-key-3 passage-offset passage-text passage-annotation-id-1 ... passage-annotation-location-offset-3 passage-annotation-location-offset-4 passage-annotation-location-length-1 passage-annotation-location-length-2 passage-annotation-location-length-3 passage-annotation-location-length-4 passage-annotation-text-1 passage-annotation-text-2 passage-annotation-text-3 passage-annotation-text-4
0 NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
1 NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
2 NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN ... NaN NaN NaN NaN NaN NaN NaN NaN NaN NaN
3 2301222206 no 1 1 tt_curatable tt_version tt_round 0 Johnny likes pizza and chocolate, he lives in Italy with Emily. 1 ... 46 57 6 19 5 5 Johnny pizza and chocolate Italy Emily
4 rows × 56 columns
- with same question, first answer using pandas_read_xml library
import pandas_read_xml as pdx
p2 = 'Johnny.xml'
df = pdx.read_xml(p2, ['collection'])
df = pdx.fully_flatten(df)
df
result generated 47 rows, again was not what I was looking for.
- also tried with beautifulsoup as suggested here
Thank you!
CodePudding user response:
Here's an example using pandas and xmltodict
import pandas as pd
import xmltodict
from pathlib import Path
xmldict = xmltodict.parse(Path("johnny.xml").read_text())
# unpack the names/text content from document.infon
xmldict["collection"]["document"]["infon"] = dict(
list(row.values())
for row in xmldict["collection"]["document"]["infon"]
)
# unpack the names/text content from annotation dicts
xmldict["collection"]["document"]["passage"]["annotation"] = [
{ key: val for key, val in row.items() if key != "infon" } |
{ col["@key"]: col.get("#text") for col in row["infon"] }
for row in xmldict["collection"]["document"]["passage"]["annotation"]
]
# use `.json_normalize()` to create a dataframe
# `.explode()` turns each annotation into its own row
df = (
pd.json_normalize(xmldict)
.explode("collection.document.passage.annotation")
)
# remove annotations column
# use `.json_normalize()` to create dataframe from annotation dicts
# concat/combine the columns with original dataframe
df = pd.concat(
[
df.drop(columns="collection.document.passage.annotation"),
pd.json_normalize(df["collection.document.passage.annotation"])
.set_index(df.index)
],
axis=1
)
You can rename/remove columns as desired:
>>> df.columns
Index(['collection.source', 'collection.date', 'collection.key',
'collection.document.id', 'collection.document.infon.tt_curatable',
'collection.document.infon.tt_version',
'collection.document.infon.tt_round',
'collection.document.passage.offset',
'collection.document.passage.text',
'@id', 'text', 'type', 'identifier', 'annotator', 'updated_at',
'location.@offset', 'location.@length'],
dtype='object')
>>> df[["@id", "text", "type", "identifier"]]
@id text type identifier
0 1 Johnny names first_name
0 3 pizza and chocolate food None
0 4 Italy location europe
0 2 Emily names first_name
[UPDATE]:
Possible alternative approach with the use of |
for row in xmldict["collection"]["document"]["passage"]["annotation"]:
row.update(
{ col["@key"]: col.get("#text") for col in row["infon"] }
)
row.pop("infon", None)
What happens is row goes from:
{'@id': '1',
'infon': [
{'@key': 'type', '#text': 'names'},
{'@key': 'identifier', '#text': 'first_name'},
{'@key': 'annotator', '#text': 'annotator_1'},
{'@key': 'updated_at', '#text': '2023-01-22T22:12:56Z'}],
'location': {'@offset': '0', '@length': '6'},
'text': 'Johnny'}
Into:
{'@id': '1',
'type': 'names',
'identifier': 'first_name',
'annotator': 'annotator_1',
'updated_at': '2023-01-22T22:12:56Z',
'location': {'@offset': '0', '@length': '6'},
'text': 'Johnny'}
Each dict inside row["infon"] is "unpacked" the the key/text values are "merged" into the top-level.
The infon key is then removed.
The reason xmltodict uses @key/#text is to avoid name-clashes.
If there was an inner {"@key": "text", ...} in this example, merging it into the top-level would overwrite the existing "text": "Johnny"
If this is a concern you could prepend annotation. to the keys so you instead end up with:
{'@id': '1',
'annotation.type': 'names',
'annotation.identifier': 'first_name',
'annoation.annotator': 'annotator_1',
'annotation.updated_at': '2023-01-22T22:12:56Z',
'location': {'@offset': '0', '@length': '6'},
'text': 'Johnny'}
Which is probably what I should have done in the initial example.